Recognition of duplex RNA by the deaminase domain of the RNA editing enzyme ADAR2
- PMID: 25564529
- PMCID: PMC4333395
- DOI: 10.1093/nar/gku1345
Recognition of duplex RNA by the deaminase domain of the RNA editing enzyme ADAR2
Abstract
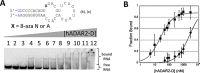
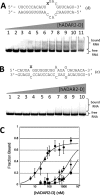
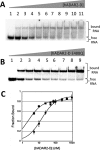
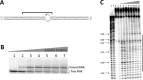
Adenosine deaminases acting on RNA (ADARs) hydrolytically deaminate adenosines (A) in a wide variety of duplex RNAs and misregulation of editing is correlated with human disease. However, our understanding of reaction selectivity is limited. ADARs are modular enzymes with multiple double-stranded RNA binding domains (dsRBDs) and a catalytic domain. While dsRBD binding is understood, little is known about ADAR catalytic domain/RNA interactions. Here we use a recently discovered RNA substrate that is rapidly deaminated by the isolated human ADAR2 deaminase domain (hADAR2-D) to probe these interactions. We introduced the nucleoside analog 8-azanebularine (8-azaN) into this RNA (and derived constructs) to mechanistically trap the protein-RNA complex without catalytic turnover for EMSA and ribonuclease footprinting analyses. EMSA showed that hADAR2-D requires duplex RNA and is sensitive to 2'-deoxy substitution at nucleotides opposite the editing site, the local sequence and 8-azaN nucleotide positioning on the duplex. Ribonuclease V1 footprinting shows that hADAR2-D protects ∼ 23 nt on the edited strand around the editing site in an asymmetric fashion (∼ 18 nt on the 5' side and ∼ 5 nt on the 3' side). These studies provide a deeper understanding of the ADAR catalytic domain-RNA interaction and new tools for biophysical analysis of ADAR-RNA complexes.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



References
-
- Burns C.M., Chu H., Rueter S.M., Hutchinson L.K., Canton H., Sanders-Bush E., Emeson R.B. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature. 1997;387:303–308. - PubMed
-
- Higuchi M., Single F.N., Kohler M., Sommer B., Sprengel R., Seeburg P.H. RNA editing of AMPA receptor subunit GluR-B: a base-paired intron-exon structure determines position and efficiency. Cell. 1993;75:1361–1370. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

