Global coordination in adaptation to gene rewiring
- PMID: 25564530
- PMCID: PMC4333410
- DOI: 10.1093/nar/gku1366
Global coordination in adaptation to gene rewiring
Abstract
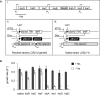
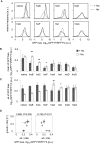
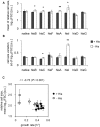
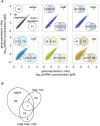
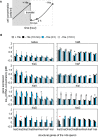
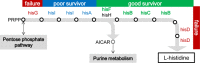
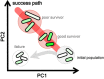
Gene rewiring is a common evolutionary phenomenon in nature that may lead to extinction for living organisms. Recent studies on synthetic biology demonstrate that cells can survive genetic rewiring. This survival (adaptation) is often linked to the stochastic expression of rewired genes with random transcriptional changes. However, the probability of adaptation and the underlying common principles are not clear. We performed a systematic survey of an assortment of gene-rewired Escherichia coli strains to address these questions. Three different cell fates, designated good survivors, poor survivors and failures, were observed when the strains starved. Large fluctuations in the expression of the rewired gene were commonly observed with increasing cell size, but these changes were insufficient for adaptation. Cooperative reorganizations in the corresponding operon and genome-wide gene expression largely contributed to the final success. Transcriptome reorganizations that generally showed high-dimensional dynamic changes were restricted within a one-dimensional trajectory for adaptation to gene rewiring, indicating a general path directed toward cellular plasticity for a successful cell fate. This finding of global coordination supports a mechanism of stochastic adaptation and provides novel insights into the design and application of complex genetic or metabolic networks.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
-
- Davison E. Genomic Regulatory Systems, Development and Evolution. London: Academic Press; 2001.
-
- Wilkins A.S. The Evolution of Developmental Pathways. Sunderland, MA: Sinauer Associates; 2002.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

