A heteromeric membrane-bound prenyltransferase complex from hop catalyzes three sequential aromatic prenylations in the bitter acid pathway
- PMID: 25564559
- PMCID: PMC4348772
- DOI: 10.1104/pp.114.253682
A heteromeric membrane-bound prenyltransferase complex from hop catalyzes three sequential aromatic prenylations in the bitter acid pathway
Abstract
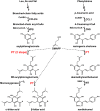
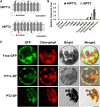
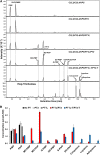
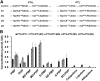
Bitter acids (α and β types) account for more than 30% of the fresh weight of hop (Humulus lupulus) glandular trichomes and are well known for their contribution to the bitter taste of beer. These multiprenylated chemicals also show diverse biological activities, some of which have potential benefits to human health. The bitter acid biosynthetic pathway has been investigated extensively, and the genes for the early steps of bitter acid synthesis have been cloned and functionally characterized. However, little is known about the enzyme(s) that catalyze three sequential prenylation steps in the β-bitter acid pathway. Here, we employed a yeast (Saccharomyces cerevisiae) system for the functional identification of aromatic prenyltransferase (PT) genes. Two PT genes (HlPT1L and HlPT2) obtained from a hop trichome-specific complementary DNA library were functionally characterized using this yeast system. Coexpression of codon-optimized PT1L and PT2 in yeast, together with upstream genes, led to the production of bitter acids, but no bitter acids were detected when either of the PT genes was expressed by itself. Stepwise mutation of the aspartate-rich motifs in PT1L and PT2 further revealed the prenylation sequence of these two enzymes in β-bitter acid biosynthesis: PT1L catalyzed only the first prenylation step, and PT2 catalyzed the two subsequent prenylation steps. A metabolon formed through interactions between PT1L and PT2 was demonstrated using a yeast two-hybrid system, reciprocal coimmunoprecipitation, and in vitro biochemical assays. These results provide direct evidence of the involvement of a functional metabolon of membrane-bound prenyltransferases in bitter acid biosynthesis in hop.
© 2015 American Society of Plant Biologists. All Rights Reserved.
Figures


Similar articles
-
HlPT-1, a membrane-bound prenyltransferase responsible for the biosynthesis of bitter acids in hops.Biochem Biophys Res Commun. 2012 Jan 6;417(1):393-8. doi: 10.1016/j.bbrc.2011.11.125. Epub 2011 Dec 7. Biochem Biophys Res Commun. 2012. PMID: 22166201
-
Characterization of the formation of branched short-chain fatty acid:CoAs for bitter acid biosynthesis in hop glandular trichomes.Mol Plant. 2013 Jul;6(4):1301-17. doi: 10.1093/mp/sst004. Epub 2013 Jan 8. Mol Plant. 2013. PMID: 23300257
-
Transcriptome analysis of bitter acid biosynthesis and precursor pathways in hop (Humulus lupulus).BMC Plant Biol. 2013 Jan 24;13:12. doi: 10.1186/1471-2229-13-12. BMC Plant Biol. 2013. PMID: 23347725 Free PMC article.
-
Prenylation of aromatic compounds, a key diversification of plant secondary metabolites.Phytochemistry. 2009 Oct-Nov;70(15-16):1739-45. doi: 10.1016/j.phytochem.2009.08.023. Epub 2009 Oct 12. Phytochemistry. 2009. PMID: 19819506 Review.
-
Prenyltransferases as key enzymes in primary and secondary metabolism.Appl Microbiol Biotechnol. 2015 Sep;99(18):7379-97. doi: 10.1007/s00253-015-6811-y. Epub 2015 Jul 28. Appl Microbiol Biotechnol. 2015. PMID: 26216239 Review.
Cited by
-
Convergent evolution of the UbiA prenyltransferase family underlies the independent acquisition of furanocoumarins in plants.New Phytol. 2020 Mar;225(5):2166-2182. doi: 10.1111/nph.16277. Epub 2019 Nov 19. New Phytol. 2020. PMID: 31642055 Free PMC article.
-
Isolation of Artemisia capillaris membrane-bound di-prenyltransferase for phenylpropanoids and redesign of artepillin C in yeast.Commun Biol. 2019 Oct 18;2:384. doi: 10.1038/s42003-019-0630-0. eCollection 2019. Commun Biol. 2019. PMID: 31646187 Free PMC article.
-
Hops (Humulus lupulus L.) Bitter Acids: Modulation of Rumen Fermentation and Potential As an Alternative Growth Promoter.Front Vet Sci. 2017 Aug 21;4:131. doi: 10.3389/fvets.2017.00131. eCollection 2017. Front Vet Sci. 2017. PMID: 28871284 Free PMC article. Review.
-
Harvesting the biosynthetic machineries that cultivate a variety of indispensable plant natural products.Curr Opin Chem Biol. 2016 Apr;31:66-73. doi: 10.1016/j.cbpa.2016.01.008. Epub 2016 Feb 4. Curr Opin Chem Biol. 2016. PMID: 26851514 Free PMC article. Review.
-
Dearomative gem-diprenylation of hydroxynaphthalenes by an engineered fungal prenyltransferase.RSC Adv. 2022 Sep 27;12(42):27550-27554. doi: 10.1039/d2ra04837j. eCollection 2022 Sep 22. RSC Adv. 2022. PMID: 36276050 Free PMC article.
References
-
- Caballero I, Blanco CA, Porras M (2012) Iso-alpha-acids, bitterness and loss of beer quality during storage. Trends Food Sci Technol 26: 21–30
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

