Model-driven discovery of underground metabolic functions in Escherichia coli
- PMID: 25564669
- PMCID: PMC4311852
- DOI: 10.1073/pnas.1414218112
Model-driven discovery of underground metabolic functions in Escherichia coli
Abstract
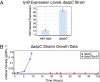
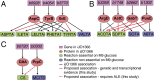
Enzyme promiscuity toward substrates has been discussed in evolutionary terms as providing the flexibility to adapt to novel environments. In the present work, we describe an approach toward exploring such enzyme promiscuity in the space of a metabolic network. This approach leverages genome-scale models, which have been widely used for predicting growth phenotypes in various environments or following a genetic perturbation; however, these predictions occasionally fail. Failed predictions of gene essentiality offer an opportunity for targeting biological discovery, suggesting the presence of unknown underground pathways stemming from enzymatic cross-reactivity. We demonstrate a workflow that couples constraint-based modeling and bioinformatic tools with KO strain analysis and adaptive laboratory evolution for the purpose of predicting promiscuity at the genome scale. Three cases of genes that are incorrectly predicted as essential in Escherichia coli--aspC, argD, and gltA--are examined, and isozyme functions are uncovered for each to a different extent. Seven isozyme functions based on genetic and transcriptional evidence are suggested between the genes aspC and tyrB, argD and astC, gabT and puuE, and gltA and prpC. This study demonstrates how a targeted model-driven approach to discovery can systematically fill knowledge gaps, characterize underground metabolism, and elucidate regulatory mechanisms of adaptation in response to gene KO perturbations.
Keywords: genome-scale modeling; isozyme discovery; substrate promiscuity; systems biology; underground metabolism.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Enzyme promiscuity shapes adaptation to novel growth substrates.Mol Syst Biol. 2019 Apr 8;15(4):e8462. doi: 10.15252/msb.20188462. Mol Syst Biol. 2019. PMID: 30962359 Free PMC article.
-
Reframing gene essentiality in terms of adaptive flexibility.BMC Syst Biol. 2018 Dec 17;12(1):143. doi: 10.1186/s12918-018-0653-z. BMC Syst Biol. 2018. PMID: 30558585 Free PMC article.
-
Network-level architecture and the evolutionary potential of underground metabolism.Proc Natl Acad Sci U S A. 2014 Aug 12;111(32):11762-7. doi: 10.1073/pnas.1406102111. Epub 2014 Jul 28. Proc Natl Acad Sci U S A. 2014. PMID: 25071190 Free PMC article.
-
How enzyme promiscuity and horizontal gene transfer contribute to metabolic innovation.FEBS J. 2020 Apr;287(7):1323-1342. doi: 10.1111/febs.15185. Epub 2020 Jan 10. FEBS J. 2020. PMID: 31858709 Free PMC article. Review.
-
Systems-level modeling of mycobacterial metabolism for the identification of new (multi-)drug targets.Semin Immunol. 2014 Dec;26(6):610-22. doi: 10.1016/j.smim.2014.09.013. Epub 2014 Oct 23. Semin Immunol. 2014. PMID: 25453232 Review.
Cited by
-
Filling gaps in metabolism using hypothetical reactions.Proc Natl Acad Sci U S A. 2022 Dec 6;119(49):e2217400119. doi: 10.1073/pnas.2217400119. Epub 2022 Nov 29. Proc Natl Acad Sci U S A. 2022. PMID: 36442125 Free PMC article. No abstract available.
-
Gradients in gene essentiality reshape antibacterial research.FEMS Microbiol Rev. 2022 May 6;46(3):fuac005. doi: 10.1093/femsre/fuac005. FEMS Microbiol Rev. 2022. PMID: 35104846 Free PMC article. Review.
-
Enzyme promiscuity shapes adaptation to novel growth substrates.Mol Syst Biol. 2019 Apr 8;15(4):e8462. doi: 10.15252/msb.20188462. Mol Syst Biol. 2019. PMID: 30962359 Free PMC article.
-
Reconstruction and metabolic profiling of the genome-scale metabolic network model of Pseudomonas stutzeri A1501.Synth Syst Biotechnol. 2023 Oct 20;8(4):688-696. doi: 10.1016/j.synbio.2023.10.001. eCollection 2023 Dec. Synth Syst Biotechnol. 2023. PMID: 37927897 Free PMC article.
-
Computation-aided designs enable developing auxotrophic metabolic sensors for wide-range glyoxylate and glycolate detection.Nat Commun. 2025 Mar 4;16(1):2168. doi: 10.1038/s41467-025-57407-3. Nat Commun. 2025. PMID: 40038270 Free PMC article.
References
-
- Tawfik DS. Accuracy-rate tradeoffs: How do enzymes meet demands of selectivity and catalytic efficiency? Curr Opin Chem Biol. 2014;21:73–80. - PubMed
-
- Jensen RA. Enzyme recruitment in evolution of new function. Annu Rev Microbiol. 1976;30:409–425. - PubMed
-
- Lazcano A, Miller SL. The origin and early evolution of life: Prebiotic chemistry, the pre-RNA world, and time. Cell. 1996;85(6):793–798. - PubMed
-
- Rison SCG, Thornton JM. Pathway evolution, structurally speaking. Curr Opin Struct Biol. 2002;12(3):374–382. - PubMed
-
- Khersonsky O, Tawfik DS. Enzyme promiscuity: A mechanistic and evolutionary perspective. Annu Rev Biochem. 2010;79:471–505. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

