TBX6 null variants and a common hypomorphic allele in congenital scoliosis
- PMID: 25564734
- PMCID: PMC4326244
- DOI: 10.1056/NEJMoa1406829
TBX6 null variants and a common hypomorphic allele in congenital scoliosis
Abstract
Background: Congenital scoliosis is a common type of vertebral malformation. Genetic susceptibility has been implicated in congenital scoliosis.
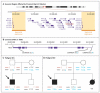
Methods: We evaluated 161 Han Chinese persons with sporadic congenital scoliosis, 166 Han Chinese controls, and 2 pedigrees, family members of which had a 16p11.2 deletion, using comparative genomic hybridization, quantitative polymerase-chain-reaction analysis, and DNA sequencing. We carried out tests of replication using an additional series of 76 Han Chinese persons with congenital scoliosis and a multicenter series of 42 persons with 16p11.2 deletions.
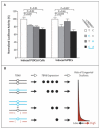
Results: We identified a total of 17 heterozygous TBX6 null mutations in the 161 persons with sporadic congenital scoliosis (11%); we did not observe any null mutations in TBX6 in 166 controls (P<3.8×10(-6)). These null alleles include copy-number variants (12 instances of a 16p11.2 deletion affecting TBX6) and single-nucleotide variants (1 nonsense and 4 frame-shift mutations). However, the discordant intrafamilial phenotypes of 16p11.2 deletion carriers suggest that heterozygous TBX6 null mutation is insufficient to cause congenital scoliosis. We went on to identify a common TBX6 haplotype as the second risk allele in all 17 carriers of TBX6 null mutations (P<1.1×10(-6)). Replication studies involving additional persons with congenital scoliosis who carried a deletion affecting TBX6 confirmed this compound inheritance model. In vitro functional assays suggested that the risk haplotype is a hypomorphic allele. Hemivertebrae are characteristic of TBX6-associated congenital scoliosis.
Conclusions: Compound inheritance of a rare null mutation and a hypomorphic allele of TBX6 accounted for up to 11% of congenital scoliosis cases in the series that we analyzed. (Funded by the National Basic Research Program of China and others.).
Figures

References
-
- Giampietro PF, Dunwoodie SL, Kusumi K, et al. Progress in the understanding of the genetic etiology of vertebral segmentation disorders in humans. Ann N Y Acad Sci. 2009;1151:38–67. - PubMed
-
- Sparrow DB, Chapman G, Smith AJ, et al. A mechanism for gene-environment interaction in the etiology of congenital scoliosis. Cell. 2012;149:295–306. - PubMed
-
- Ghebranious N, Blank RD, Raggio CL, et al. A missense T (Brachyury) mutation contributes to vertebral malformations. J Bone Miner Res. 2008;23:1576–83. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
