Synonymous mutations reduce genome compactness in icosahedral ssRNA viruses
- PMID: 25564866
- PMCID: PMC4286596
- DOI: 10.1016/j.bpj.2014.10.070
Synonymous mutations reduce genome compactness in icosahedral ssRNA viruses
Abstract
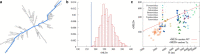
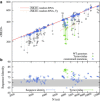
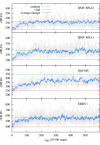
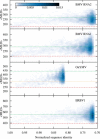
Recent studies have shown that single-stranded (ss) viral RNAs fold into more compact structures than random RNA sequences with similar chemical composition and identical length. Based on this comparison, it has been suggested that wild-type viral RNA may have evolved to be atypically compact so as to aid its encapsidation and assist the viral assembly process. To further explore the compactness selection hypothesis, we systematically compare the predicted sizes of >100 wild-type viral sequences with those of their mutants, which are evolved in silico and subject to a number of known evolutionary constraints. In particular, we enforce mutation synonynimity, preserve the codon-bias, and leave untranslated regions intact. It is found that progressive accumulation of these restricted mutations still suffices to completely erase the characteristic compactness imprint of the viral RNA genomes, making them in this respect physically indistinguishable from randomly shuffled RNAs. This shows that maintaining the physical compactness of the genome is indeed a primary factor among ssRNA viruses' evolutionary constraints, contributing also to the evidence that synonymous mutations in viral ssRNA genomes are not strictly neutral.
Copyright © 2015 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures
Comment in
-
Viral ssRNAs are indeed compact.Biophys J. 2015 Jan 6;108(1):14-6. doi: 10.1016/j.bpj.2014.11.010. Biophys J. 2015. PMID: 25564845 Free PMC article. No abstract available.
References
-
- Dykeman E.C., Stockley P.G., Twarock R. Packaging signals in two single-stranded RNA viruses imply a conserved assembly mechanism and geometry of the packaged genome. J. Mol. Biol. 2013;425:3235–3249. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

