From guide to target: molecular insights into eukaryotic RNA-interference machinery
- PMID: 25565029
- PMCID: PMC4450863
- DOI: 10.1038/nsmb.2931
From guide to target: molecular insights into eukaryotic RNA-interference machinery
Abstract
Since its relatively recent discovery, RNA interference (RNAi) has emerged as a potent, specific and ubiquitous means of gene regulation. Through a number of pathways that are conserved in eukaryotes from yeast to humans, small noncoding RNAs direct molecular machinery to silence gene expression. In this Review, we focus on mechanisms and structures that govern RNA silencing in higher organisms. In addition to highlighting recent advances, we discuss parallels and differences among RNAi pathways. Together, the studies reviewed herein reveal the versatility and programmability of RNA-induced silencing complexes and emphasize the importance of both upstream biogenesis and downstream silencing factors.
Figures
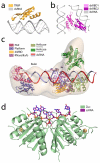
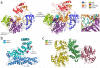
References
-
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806. - PubMed
-
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843. - PubMed
-
- Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

