Pyruvate kinase M2 regulates Hif-1α activity and IL-1β induction and is a critical determinant of the warburg effect in LPS-activated macrophages
- PMID: 25565206
- PMCID: PMC5198835
- DOI: 10.1016/j.cmet.2014.12.005
Pyruvate kinase M2 regulates Hif-1α activity and IL-1β induction and is a critical determinant of the warburg effect in LPS-activated macrophages
Erratum in
- Cell Metab. 2015 Feb 3;21(2):347. Jiang, Jain-Kang [corrected to Jiang, Jian-Kang]; Vanden Heiden, Matthew [corrected to Vander Heiden, Matthew]
-
Pyruvate Kinase M2 Regulates Hif-1α Activity and IL-1β Induction and Is a Critical Determinant of the Warburg Effect in LPS-Activated Macrophages.Cell Metab. 2015 Feb 3;21(2):347. doi: 10.1016/j.cmet.2015.01.017. Epub 2015 Feb 3. Cell Metab. 2015. PMID: 29510100 No abstract available.
Abstract
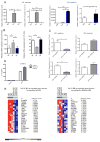
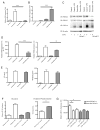
Macrophages activated by the TLR4 agonist LPS undergo dramatic changes in their metabolic activity. We here show that LPS induces expression of the key metabolic regulator Pyruvate Kinase M2 (PKM2). Activation of PKM2 using two well-characterized small molecules, DASA-58 and TEPP-46, inhibited LPS-induced Hif-1α and IL-1β, as well as the expression of a range of other Hif-1α-dependent genes. Activation of PKM2 attenuated an LPS-induced proinflammatory M1 macrophage phenotype while promoting traits typical of an M2 macrophage. We show that LPS-induced PKM2 enters into a complex with Hif-1α, which can directly bind to the IL-1β promoter, an event that is inhibited by activation of PKM2. Both compounds inhibited LPS-induced glycolytic reprogramming and succinate production. Finally, activation of PKM2 by TEPP-46 in vivo inhibited LPS and Salmonella typhimurium-induced IL-1β production, while boosting production of IL-10. PKM2 is therefore a critical determinant of macrophage activation by LPS, promoting the inflammatory response.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures





References
-
- Altenberg B, Greulich KO. Genes of glycolysis are ubiquitously overexpressed in 24 cancer classes. Genomics. 2004;84:1014–1020. - PubMed
-
- Ashizawa K, Willingham MC, Liang CM, Cheng SY. In vivo regulation of monomer-tetramer conversion of pyruvate kinase subtype M2 by glucose is mediated via fructose 1,6-bisphosphate. The Journal of biological chemistry. 1991;266:16842–16846. - PubMed
-
- Boxer MB, Jiang JK, Vander Heiden MG, Shen M, Skoumbourdis AP, Southall N, Veith H, Leister W, Austin CP, Park HW, et al. Evaluation of substituted N,N′-diarylsulfonamides as activators of the tumor cell specific M2 isoform of pyruvate kinase. Journal of medicinal chemistry. 2010;53:1048–1055. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

