Decarbamoyl mitomycin C (DMC) activates p53-independent ataxia telangiectasia and rad3 related protein (ATR) chromatin eviction
- PMID: 25565400
- PMCID: PMC4418290
- DOI: 10.1080/15384101.2014.997517
Decarbamoyl mitomycin C (DMC) activates p53-independent ataxia telangiectasia and rad3 related protein (ATR) chromatin eviction
Abstract
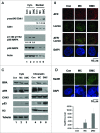
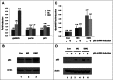
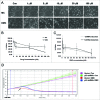
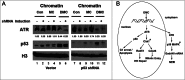
Interstrand crosslinks induce DNA replication fork stalling that in turn activates the ATR-dependent checkpoint and DNA repair on nuclear chromatin. Mitomycin C (MC) and Decarbamoyl Mitomycin C (DMC) induce different types of DNA crosslinks with DMC being a more cytotoxic agent. We previously reported that the novel DMC induced β-interstrand DNA crosslinks induce a p53-independent form of cell death. The p53-independent DMC cytotoxicity associates with the activation, and subsequent depletion, of Chk1. In this study we further dissect the novel DMC signal transduction pathway and asked how it influences chromatin-associated proteins. We found that treatment with DMC, but not MC, stimulated the disassociation of ATR from chromatin and re-localization of ATR to the cytoplasm. The chromatin eviction of ATR was coupled with the formation of nuclear Rad51 foci and the phosphorylation of Chk1. Furthermore, DMC but not MC, activated expression of gadd45α mRNA. Importantly, knocking down p53 via shRNA did not inhibit the DMC-induced disassociation of ATR from chromatin or reduce the activation of transcription of gadd45α. Our results suggest that DMC induces a p53-independent disassociation of ATR from chromatin that facilitates Chk1 checkpoint activation and Rad51 chromatin recruitment. Our findings provide evidence that ATR chromatin eviction in breast cancer cells is an area of study that should be focused on for inducing p53-independent cell death.
Keywords: ATR; ATR, Ataxia Telangiectasia and Rad3 Related; Cdc25, cell division cycle 25.; ChIP, chromatin immunoprecipitation; Chk1, checkpoint serine/threonine protein kinase 1; DDR, DNA damage response; DMC, Decarbamoyl Mitomycin C; DOX, doxycycline; HR, homologous recombination; ICLs, interstrand cross-links; MC, Mitomycin C; NER, nuclear excision repair; PCNA, Proliferating Cell Nuclear Antigen; RPA, replication protein A; TNBCs, triple negative breast cancers; breast cancer; cell death; chromatin; interstrand crosslink; mitomycin C; p53.
Figures

Similar articles
-
DNA adducts of decarbamoyl mitomycin C efficiently kill cells without wild-type p53 resulting from proteasome-mediated degradation of checkpoint protein 1.Chem Res Toxicol. 2010 Jul 19;23(7):1151-62. doi: 10.1021/tx900420k. Chem Res Toxicol. 2010. PMID: 20536192 Free PMC article.
-
Activation of the S phase DNA damage checkpoint by mitomycin C.J Cell Physiol. 2007 May;211(2):468-76. doi: 10.1002/jcp.20957. J Cell Physiol. 2007. PMID: 17167777
-
S-phase sensing of DNA-protein crosslinks triggers TopBP1-independent ATR activation and p53-mediated cell death by formaldehyde.Cell Cycle. 2012 Jul 1;11(13):2526-37. doi: 10.4161/cc.20905. Epub 2012 Jul 1. Cell Cycle. 2012. PMID: 22722496 Free PMC article.
-
Regulation of ATR-CHK1 signaling by ubiquitination of CLASPIN.Biochem Soc Trans. 2022 Oct 31;50(5):1471-1480. doi: 10.1042/BST20220729. Biochem Soc Trans. 2022. PMID: 36196914 Review.
-
Novel regulation of checkpoint kinase 1: Is checkpoint kinase 1 a good candidate for anti-cancer therapy?Cancer Sci. 2012 Jul;103(7):1195-200. doi: 10.1111/j.1349-7006.2012.02280.x. Epub 2012 Apr 23. Cancer Sci. 2012. PMID: 22435685 Free PMC article. Review.
Cited by
-
Mitomycin C and its analog trigger cytotoxicity in MCF-7 and K562 cancer cells through the regulation of RAS and MAPK/ERK pathways.Chem Biol Interact. 2024 May 25;395:111007. doi: 10.1016/j.cbi.2024.111007. Epub 2024 Apr 18. Chem Biol Interact. 2024. PMID: 38642817 Free PMC article.
-
Mitomycin C and decarbamoyl mitomycin C induce p53-independent p21WAF1/CIP1 activation.Int J Oncol. 2016 Nov;49(5):1815-1824. doi: 10.3892/ijo.2016.3703. Epub 2016 Sep 23. Int J Oncol. 2016. PMID: 27666201 Free PMC article.
-
Isolation and Rationale for the Formation of Isomeric Decarbamoylmitomycin C- N6-deoxyadenosine Adducts in DNA.Chem Res Toxicol. 2018 Aug 20;31(8):762-771. doi: 10.1021/acs.chemrestox.8b00102. Epub 2018 Aug 2. Chem Res Toxicol. 2018. PMID: 30035537 Free PMC article.
-
Synthesis of Oligonucleotides containing the cis-Interstrand Crosslink Produced by Mitomycins in their Reaction with DNA.Chemistry. 2020 Oct 1;26(55):12570-12578. doi: 10.1002/chem.202002452. Epub 2020 Sep 3. Chemistry. 2020. PMID: 32574396 Free PMC article.
-
Cytotoxicity, crosslinking and biological activity of three mitomycins.Bioorg Chem. 2022 Jun;123:105744. doi: 10.1016/j.bioorg.2022.105744. Epub 2022 Mar 22. Bioorg Chem. 2022. PMID: 35349830 Free PMC article.
References
-
- Garewal HS. (1988) Mitomycin C in the chemotherapy of advanced breast cancer, Semin Oncol 15:74-9; PMID:3134698 - PubMed
-
- Bradner WT. (2001) Mitomycin C: a clinical update, Cancer Treat Rev 27:35-50; PMID:11237776; http://dx.doi.org/10.1053/ctrv.2000.0202 - DOI - PubMed
-
- Bargonetti J, Champeil E, and Tomasz M. (2010) Differential toxicity of DNA adducts of mitomycin C, J Nucleic Acids 2010; PMID:20798760; http://dx.doi.org/10.4061/2010/698960 - DOI - PMC - PubMed
-
- Lawley PD, Phillips DH. (1996) DNA adducts from chemotherapeutic agents, Mutat Res 355:13-40; PMID:8781575; http://dx.doi.org/10.1016/0027-5107(96)00020-6 - DOI - PubMed
-
- Boamah EK, Brekman A, Tomasz M, Myeku N, Figueiredo-Pereira M, Hunter S, Meyer J, Bhosle RC, Bargonetti J. (2010) DNA adducts of decarbamoyl mitomycin C efficiently kill cells without wild-type p53 resulting from proteasome-mediated degradation of checkpoint protein 1, Chem Res Toxicol 23:1151-62; PMID:20536192; http://dx.doi.org/10.1021/tx900420k - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
