Transcript co-variance with Nestin in two mouse genetic reference populations identifies Lef1 as a novel candidate regulator of neural precursor cell proliferation in the adult hippocampus
- PMID: 25565948
- PMCID: PMC4264481
- DOI: 10.3389/fnins.2014.00418
Transcript co-variance with Nestin in two mouse genetic reference populations identifies Lef1 as a novel candidate regulator of neural precursor cell proliferation in the adult hippocampus
Abstract
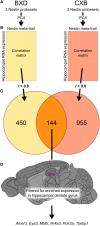
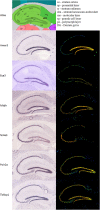
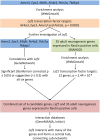
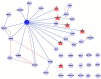
Adult neurogenesis, the lifelong production of new neurons in the adult brain, is under complex genetic control but many of the genes involved remain to be identified. In this study, we have integrated publicly available gene expression data from the BXD and CXB recombinant inbred mouse lines to discover genes co-expressed in the adult hippocampus with Nestin, a common marker of the neural precursor cell population. In addition, we incorporated spatial expression information to restrict candidates to genes with high differential gene expression in the hippocampal dentate gyrus. Incorporating data from curated protein-protein interaction databases revealed interactions between our candidate genes and those already known to be involved in adult neurogenesis. Enrichment analysis suggested a link to the Wnt/β-catenin pathway, known to be involved in adult neurogenesis. In particular, our candidates were enriched in targets of Lef1, a modulator of the Wnt pathway. In conclusion, our combination of bioinformatics approaches identified six novel candidate genes involved in adult neurogenesis; Amer3, Eya3, Mtdh, Nr4a3, Polr2a, and Tbkbp1. Further, we propose a role for Lef1 transcriptional control in the regulation of adult hippocampal precursor cell proliferation.
Keywords: BXD; CXB; Lef1; Wnt pathway; adult neurogenesis; neuroinformatics; recombinant inbred mice; systems genetics.
Figures

References
-
- Allen Institute for Brain Science (2014). Allen Mouse Brain Atlas [Internet]. Available online at: http://mouse.brain-map.org/
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

