High temporal and spatial diversity in marine RNA viruses implies that they have an important role in mortality and structuring plankton communities
- PMID: 25566218
- PMCID: PMC4266044
- DOI: 10.3389/fmicb.2014.00703
High temporal and spatial diversity in marine RNA viruses implies that they have an important role in mortality and structuring plankton communities
Abstract
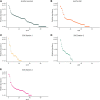
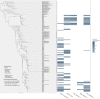
Viruses in the order Picornavirales infect eukaryotes, and are widely distributed in coastal waters. Amplicon deep-sequencing of the RNA dependent RNA polymerase (RdRp) revealed diverse and highly uneven communities of picorna-like viruses in the coastal waters of British Columbia (BC), Canada. Almost 300 000 pyrosequence reads revealed 145 operational taxonomic units (OTUs) based on 95% sequence similarity at the amino-acid level. Each sample had between 24 and 71 OTUs and there was little overlap among samples. Phylogenetic analysis revealed that some clades of OTUs were only found at one site; whereas, other clades included OTUs from all sites. Since most of these OTUs are likely from viruses that infect eukaryotic phytoplankton, and viral isolates infecting phytoplankton are strain-specific; each OTU probably arose from the lysis of a specific phytoplankton taxon. Moreover, the patchiness in OTU distribution, and the high turnover of viruses in the mixed layer, implies continuous infection and lysis by RNA viruses of a diverse array of eukaryotic phytoplankton taxa. Hence, these viruses are likely important elements structuring the phytoplankton community, and play a significant role in nutrient cycling and energy transfer.
Keywords: Picornavirales; RNA viruses; phytoplankton mortality; pyrosequencing; viral diversity; viral ecology.
Figures




References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

