ncPred: ncRNA-Disease Association Prediction through Tripartite Network-Based Inference
- PMID: 25566534
- PMCID: PMC4264506
- DOI: 10.3389/fbioe.2014.00071
ncPred: ncRNA-Disease Association Prediction through Tripartite Network-Based Inference
Abstract
Motivation: Over the past few years, experimental evidence has highlighted the role of microRNAs to human diseases. miRNAs are critical for the regulation of cellular processes, and, therefore, their aberration can be among the triggering causes of pathological phenomena. They are just one member of the large class of non-coding RNAs, which include transcribed ultra-conserved regions (T-UCRs), small nucleolar RNAs (snoRNAs), PIWI-interacting RNAs (piRNAs), large intergenic non-coding RNAs (lincRNAs) and, the heterogeneous group of long non-coding RNAs (lncRNAs). Their associations with diseases are few in number, and their reliability is questionable. In literature, there is only one recent method proposed by Yang et al. (2014) to predict lncRNA-disease associations. This technique, however, lacks in prediction quality. All these elements entail the need to investigate new bioinformatics tools for the prediction of high quality ncRNA-disease associations. Here, we propose a method called ncPred for the inference of novel ncRNA-disease association based on recommendation technique. We represent our knowledge through a tripartite network, whose nodes are ncRNAs, targets, or diseases. Interactions in such a network associate each ncRNA with a disease through its targets. Our algorithm, starting from such a network, computes weights between each ncRNA-disease pair using a multi-level resource transfer technique that at each step takes into account the resource transferred in the previous one.
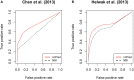
Results: The results of our experimental analysis show that our approach is able to predict more biologically significant associations with respect to those obtained by Yang et al. (2014), yielding an improvement in terms of the average area under the ROC curve (AUC). These results prove the ability of our approach to predict biologically significant associations, which could lead to a better understanding of the molecular processes involved in complex diseases.
Availability: All the ncPred predictions together with the datasets used for the analysis are available at the following url: http://alpha.dmi.unict.it/ncPred/
Keywords: lncRNAs functional characterization; ncRNAs-diseases association predictions; network-based inference; resource transfer algorithm; tripartite networks.
Figures
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

