The rebirth of culture in microbiology through the example of culturomics to study human gut microbiota
- PMID: 25567229
- PMCID: PMC4284300
- DOI: 10.1128/CMR.00014-14
The rebirth of culture in microbiology through the example of culturomics to study human gut microbiota
Abstract
Bacterial culture was the first method used to describe the human microbiota, but this method is considered outdated by many researchers. Metagenomics studies have since been applied to clinical microbiology; however, a "dark matter" of prokaryotes, which corresponds to a hole in our knowledge and includes minority bacterial populations, is not elucidated by these studies. By replicating the natural environment, environmental microbiologists were the first to reduce the "great plate count anomaly," which corresponds to the difference between microscopic and culture counts. The revolution in bacterial identification also allowed rapid progress. 16S rRNA bacterial identification allowed the accurate identification of new species. Mass spectrometry allowed the high-throughput identification of rare species and the detection of new species. By using these methods and by increasing the number of culture conditions, culturomics allowed the extension of the known human gut repertoire to levels equivalent to those of pyrosequencing. Finally, taxonogenomics strategies became an emerging method for describing new species, associating the genome sequence of the bacteria systematically. We provide a comprehensive review on these topics, demonstrating that both empirical and hypothesis-driven approaches will enable a rapid increase in the identification of the human prokaryote repertoire.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures


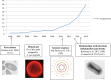
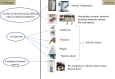
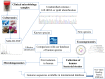






References
-
- Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, Wu D, Paulsen I, Nelson KE, Nelson W, Fouts DE, Levy S, Knap AH, Lomas MW, Nealson K, White O, Peterson J, Hoffman J, Parsons R, Baden-Tillson H, Pfannkoch C, Rogers YH, Smith HO. 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304:66–74. doi: 10.1126/science.1093857. - DOI - PubMed
-
- Lagier JC, Armougom F, Million M, Hugon P, Pagnier I, Robert C, Bittar F, Fournous G, Gimenez G, Maraninchi M, Trape JF, Koonin EV, La Scola B, Raoult D. 2012. Microbial culturomics: paradigm shift in the human gut microbiome study. Clin Microbiol Infect 18:1185–1193. doi: 10.1111/1469-0691.12023. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

