A composite enhancer regulates p63 gene expression in epidermal morphogenesis and in keratinocyte differentiation by multiple mechanisms
- PMID: 25567987
- PMCID: PMC4333422
- DOI: 10.1093/nar/gku1396
A composite enhancer regulates p63 gene expression in epidermal morphogenesis and in keratinocyte differentiation by multiple mechanisms
Abstract
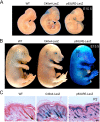
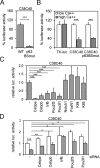
p63 is a crucial regulator of epidermal development, but its transcriptional control has remained elusive. Here, we report the identification of a long-range enhancer (p63LRE) that is composed of two evolutionary conserved modules (C38 and C40), acting in concert to control tissue- and layer-specific expression of the p63 gene. Both modules are in an open and active chromatin state in human and mouse keratinocytes and in embryonic epidermis, and are strongly bound by p63. p63LRE activity is dependent on p63 expression in embryonic skin, and also in the commitment of human induced pluripotent stem cells toward an epithelial cell fate. A search for other transcription factors involved in p63LRE regulation revealed that the CAAT enhancer binding proteins Cebpa and Cebpb and the POU domain-containing protein Pou3f1 repress p63 expression during keratinocyte differentiation by binding the p63LRE enhancer. Collectively, our data indicate that p63LRE is composed of additive and partly redundant enhancer modules that act to direct robust p63 expression selectively in the basal layer of the epidermis.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures




References
-
- Mills A.A., Zheng B., Wang X.J., Vogel H., Roop D.R., Bradley A. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 1999;398:708–713. - PubMed
-
- Yang A., Schweitzer R., Sun D., Kaghad M., Walker N., Bronson R.T., Tabin C., Sharpe A., Caput D., Crum C., et al. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398:714–718. - PubMed
-
- Senoo M., Pinto F., Crum C.P., McKeon F. p63 Is essential for the proliferative potential of stem cells in stratified epithelia. Cell. 2007;129:523–536. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

