Brg1-dependent chromatin remodelling is not essentially required during oligodendroglial differentiation
- PMID: 25568100
- PMCID: PMC6605249
- DOI: 10.1523/JNEUROSCI.1468-14.2015
Brg1-dependent chromatin remodelling is not essentially required during oligodendroglial differentiation
Abstract
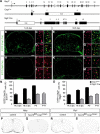
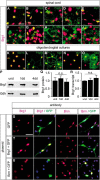
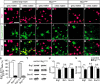
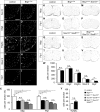
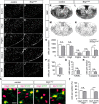
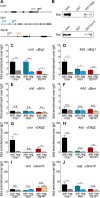
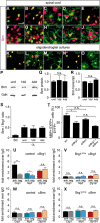
Myelinating Schwann cells in the vertebrate peripheral nervous system rely on Brg1 (Smarca4) for terminal differentiation. Brg1 serves as central ATP-hydrolyzing subunit of the chromatin remodelling BAF complexes and is recruited during myelination as part of these complexes by the transcription factor Sox10 in Schwann cells. Here, we analyzed the role of Brg1 during development of myelinating oligodendrocytes in the CNS of the mouse. Following Brg1 deletion in oligodendrocyte precursors, these cells showed normal survival, proliferation, and migration. A mild but significant reduction in the number of oligodendrocytes with myelin gene expression in the absence of Brg1 points to a contribution to oligodendroglial differentiation but also shows that the role of Brg1 is much less prominent than during Schwann cell differentiation. Additionally, we failed to obtain evidence for a genetic interaction between Brg1 and Sox10 comparable with the one in Schwann cells. This argues that similarities exist between the regulatory networks and mechanisms in both types of myelinating glia but that the exact mode of action and the relevance of functional interactions differ, pointing to a surprising degree of variability in the control of myelination.
Keywords: Schwann cell; Sox; chromatin; myelination; oligodendrocyte; transcription.
Copyright © 2015 the authors 0270-6474/15/350021-15$15.00/0.
Figures
References
-
- Ahn K, Mishina Y, Hanks MC, Behringer RR, Crenshaw EB., 3rd BMPR-IA signaling is required for the formation of the apical ectodermal ridge and dorsal-ventral patterning of the limb. Development. 2001;128:4449–4461. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
