Impact of the adenoviral E4 Orf3 protein on the activity and posttranslational modification of p53
- PMID: 25568206
- PMCID: PMC4337557
- DOI: 10.1128/JVI.03072-14
Impact of the adenoviral E4 Orf3 protein on the activity and posttranslational modification of p53
Abstract
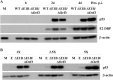
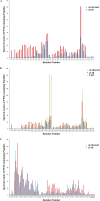
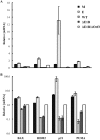
Our previous studies have established that the p53 populations that accumulate in normal human cells exposed to etoposide or infected by an E1B 55-kDa protein-null mutant of human adenovirus type 5 carry a large number of posttranslational modifications at numerous residues (C. J. DeHart, J. S. Chahal, S. J. Flint, and D. H. Perlman, Mol Cell Proteomics 13:1-17, 2014, http://dx.doi.org/10.1074/mcp.M113.030254). In the absence of this E1B protein, the p53 transcriptional program is not induced, and it has been reported that the viral E4 Orf3 protein inactivates p53 (C. Soria, F. E. Estermann, K. C. Espantman, and C. C. O'Shea, Nature 466:1076-1081, 2010, http://dx.doi.org/10.1038/nature09307). As the latter protein disrupts nuclear Pml bodies, sites at which p53 is modified, we used mass spectrometry to catalogue the posttranscriptional modifications of the p53 population that accumulates when neither the E1B 55-kDa nor the E4 Orf3 protein is made in infected cells. Eighty-five residues carrying 163 modifications were identified. The overall patterns of posttranslational modification of this population and p53 present in cells infected by an E1B 55-kDa-null mutant were similar. The efficiencies with which the two forms of p53 bound to a consensus DNA recognition sequence could not be distinguished and were lower than that of transcriptionally active p53. The absence of the E4 Orf3 protein increased expression of several p53-responsive genes when the E1B protein was also absent from infected cells. However, expression of these genes did not attain the levels observed when p53 was activated in response to etoposide treatment and remained lower than those measured in mock-infected cells.
Importance: The tumor suppressor p53, a master regulator of cellular responses to stress, is inactivated and destroyed in cells infected by species C human adenoviruses, such as type 5. It is targeted for proteasomal degradation by the action of a virus-specific E3 ubiquitin ligase that contains the viral E1B 55-kDa and E4 Orf6 proteins, while the E4 Orf3 protein has been reported to block its ability to stimulate expression of p53-dependent genes. The comparisons reported here of the posttranslational modifications and activities of p53 populations that accumulate in infected normal human cells in the absence of both mechanisms of inactivation or of only the E3 ligase revealed little impact of the E4 Orf3 protein. These observations indicate that E4 Orf3-dependent disruption of Pml bodies does not have a major effect on the pattern of p53 posttranslational modifications in adenovirus-infected cells. Furthermore, they suggest that one or more additional viral proteins contribute to blocking p53 activation and the consequences that are deleterious for viral reproduction, such as apoptosis or cell cycle arrest.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

