The interface between hepatitis B virus capsid proteins affects self-assembly, pregenomic RNA packaging, and reverse transcription
- PMID: 25568211
- PMCID: PMC4337549
- DOI: 10.1128/JVI.03545-14
The interface between hepatitis B virus capsid proteins affects self-assembly, pregenomic RNA packaging, and reverse transcription
Abstract
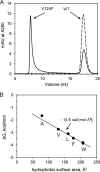
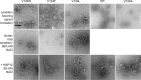
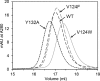
Hepatitis B virus (HBV) capsid proteins (Cps) assemble around the pregenomic RNA (pgRNA) and viral reverse transcriptase (P). pgRNA is then reverse transcribed to double-stranded DNA (dsDNA) within the capsid. The Cp assembly domain, which forms the shell of the capsid, regulates assembly kinetics and capsid stability. The Cp, via its nucleic acid-binding C-terminal domain, also affects nucleic acid organization. We hypothesize that the structure of the capsid may also have a direct effect on nucleic acid processing. Using structure-guided design, we made a series of mutations at the interface between Cp subunits that change capsid assembly kinetics and thermodynamics in a predictable manner. Assembly in cell culture mirrored in vitro activity. However, all of these mutations led to defects in pgRNA packaging. The amount of first-strand DNA synthesized was roughly proportional to the amount of RNA packaged. However, the synthesis of second-strand DNA, which requires two template switches, was not supported by any of the substitutions. These data demonstrate that the HBV capsid is far more than an inert container, as mutations in the assembly domain, distant from packaged nucleic acid, affect reverse transcription. We suggest that capsid molecular motion plays a role in regulating genome replication.
Importance: The hepatitis B virus (HBV) capsid plays a central role in the virus life cycle and has been studied as a potential antiviral target. The capsid protein (Cp) packages the viral pregenomic RNA (pgRNA) and polymerase to form the HBV core. The role of the capsid in subsequent nucleic acid metabolism is unknown. Here, guided by the structure of the capsid with bound antiviral molecules, we designed Cp mutants that enhanced or attenuated the assembly of purified Cp in vitro. In cell culture, assembly of mutants was consistent with their in vitro biophysical properties. However, all of these mutations inhibited HBV replication. Specifically, changing the biophysical chemistry of Cp caused defects in pgRNA packaging and synthesis of the second strand of DNA. These results suggest that the HBV Cp assembly domain potentially regulates reverse transcription, extending the activities of the capsid protein beyond its presumed role as an inert compartment.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures
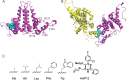


References
-
- WHO. July2004. Hepatitis B. Fact sheet no. 204. http://www.who.int/mediacentre/factsheets/fs204/en/.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

