Phosphorylation of TET proteins is regulated via O-GlcNAcylation by the O-linked N-acetylglucosamine transferase (OGT)
- PMID: 25568311
- PMCID: PMC4335217
- DOI: 10.1074/jbc.M114.605881
Phosphorylation of TET proteins is regulated via O-GlcNAcylation by the O-linked N-acetylglucosamine transferase (OGT)
Abstract
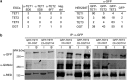
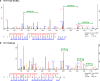
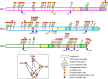
TET proteins oxidize 5-methylcytosine to 5-hydroxymethylcytosine, 5-formylcytosine, and 5-carboxylcytosine and thus provide a possible means for active DNA demethylation in mammals. Although their catalytic mechanism is well characterized and the catalytic dioxygenase domain is highly conserved, the function of the regulatory regions (the N terminus and the low-complexity insert between the two parts of the dioxygenase domains) is only poorly understood. Here, we demonstrate that TET proteins are subject to a variety of post-translational modifications that mostly occur at these regulatory regions. We mapped TET modification sites at amino acid resolution and show for the first time that TET1, TET2, and TET3 are highly phosphorylated. The O-linked GlcNAc transferase, which we identified as a strong interactor with all three TET proteins, catalyzes the addition of a GlcNAc group to serine and threonine residues of TET proteins and thereby decreases both the number of phosphorylation sites and site occupancy. Interestingly, the different TET proteins display unique post-translational modification patterns, and some modifications occur in distinct combinations. In summary, our results provide a novel potential mechanism for TET protein regulation based on a dynamic interplay of phosphorylation and O-GlcNAcylation at the N terminus and the low-complexity insert region. Our data suggest strong cross-talk between the modification sites that could allow rapid adaption of TET protein localization, activity, or targeting due to changing environmental conditions as well as in response to external stimuli.
Keywords: 5-Hydroxymethylcytosine (5-hmC); Dioxygenase; Epigenetics; O-Linked N-Acetylglucosamine (O-GlcNAc); OGT; Phosphorylation; Post-translational Modification (PTM); TET Proteins.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

