PRDM9 drives evolutionary erosion of hotspots in Mus musculus through haplotype-specific initiation of meiotic recombination
- PMID: 25568937
- PMCID: PMC4287450
- DOI: 10.1371/journal.pgen.1004916
PRDM9 drives evolutionary erosion of hotspots in Mus musculus through haplotype-specific initiation of meiotic recombination
Abstract
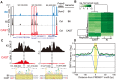
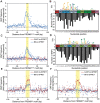
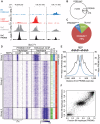
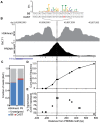
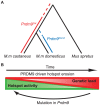
Meiotic recombination generates new genetic variation and assures the proper segregation of chromosomes in gametes. PRDM9, a zinc finger protein with histone methyltransferase activity, initiates meiotic recombination by binding DNA at recombination hotspots and directing the position of DNA double-strand breaks (DSB). The DSB repair mechanism suggests that hotspots should eventually self-destruct, yet genome-wide recombination levels remain constant, a conundrum known as the hotspot paradox. To test if PRDM9 drives this evolutionary erosion, we measured activity of the Prdm9Cst allele in two Mus musculus subspecies, M.m. castaneus, in which Prdm9Cst arose, and M.m. domesticus, into which Prdm9Cst was introduced experimentally. Comparing these two strains, we find that haplotype differences at hotspots lead to qualitative and quantitative changes in PRDM9 binding and activity. Using Mus spretus as an outlier, we found most variants affecting PRDM9Cst binding arose and were fixed in M.m. castaneus, suppressing hotspot activity. Furthermore, M.m. castaneus×M.m. domesticus F1 hybrids exhibit novel hotspots, with large haplotype biases in both PRDM9 binding and chromatin modification. These novel hotspots represent sites of historic evolutionary erosion that become activated in hybrids due to crosstalk between one parent's Prdm9 allele and the opposite parent's chromosome. Together these data support a model where haplotype-specific PRDM9 binding directs biased gene conversion at hotspots, ultimately leading to hotspot erosion.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Baudat F, Imai Y, de Massy B (2013) Meiotic recombination in mammals: localization and regulation. Nat Rev Genet 14: 794–806. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P01 GM099640/GM/NIGMS NIH HHS/United States
- R01 GM078452/GM/NIGMS NIH HHS/United States
- R01 GM078643/GM/NIGMS NIH HHS/United States
- P30 CA034196/CA/NCI NIH HHS/United States
- GM078643/GM/NIGMS NIH HHS/United States
- GM078452/GM/NIGMS NIH HHS/United States
- CA34196/CA/NCI NIH HHS/United States
- P50 GM076468/GM/NIGMS NIH HHS/United States
- T32 HD007065-32/HD/NICHD NIH HHS/United States
- T32 HD007065/HD/NICHD NIH HHS/United States
- F32 GM101736/GM/NIGMS NIH HHS/United States
- GM083408/GM/NIGMS NIH HHS/United States
- R01 GM083408/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

