Trans-splicing improvement by the combined application of antisense strategies
- PMID: 25569093
- PMCID: PMC4307297
- DOI: 10.3390/ijms16011179
Trans-splicing improvement by the combined application of antisense strategies
Abstract
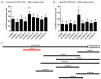
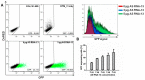
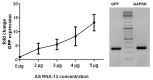
Spliceosome-mediated RNA trans-splicing has become an emergent tool for the repair of mutated pre-mRNAs in the treatment of genetic diseases. RNA trans-splicing molecules (RTMs) are designed to induce a specific trans-splicing reaction via a binding domain for a respective target pre-mRNA region. A previously established reporter-based screening system allows us to analyze the impact of various factors on the RTM trans-splicing efficiency in vitro. Using this system, we are further able to investigate the potential of antisense RNAs (AS RNAs), presuming to improve the trans-splicing efficiency of a selected RTM, specific for intron 102 of COL7A1. Mutations in the COL7A1 gene underlie the dystrophic subtype of the skin blistering disease epidermolysis bullosa (DEB). We have shown that co-transfections of the RTM and a selected AS RNA, interfering with competitive splicing elements on a COL7A1-minigene (COL7A1-MG), lead to a significant increase of the RNA trans-splicing efficiency. Thereby, accurate trans-splicing between the RTM and the COL7A1-MG is represented by the restoration of full-length green fluorescent protein GFP on mRNA and protein level. This mechanism can be crucial for the improvement of an RTM-mediated correction, especially in cases where a high trans-splicing efficiency is required.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

