Telomerase reverse transcriptase regulates microRNAs
- PMID: 25569094
- PMCID: PMC4307298
- DOI: 10.3390/ijms16011192
Telomerase reverse transcriptase regulates microRNAs
Abstract
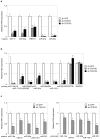
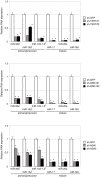
MicroRNAs are small non-coding RNAs that inhibit the translation of target mRNAs. In humans, most microRNAs are transcribed by RNA polymerase II as long primary transcripts and processed by sequential cleavage of the two RNase III enzymes, DROSHA and DICER, into precursor and mature microRNAs, respectively. Although the fundamental functions of microRNAs in RNA silencing have been gradually uncovered, less is known about the regulatory mechanisms of microRNA expression. Here, we report that telomerase reverse transcriptase (TERT) extensively affects the expression levels of mature microRNAs. Deep sequencing-based screens of short RNA populations revealed that the suppression of TERT resulted in the downregulation of microRNAs expressed in THP-1 cells and HeLa cells. Primary and precursor microRNA levels were also reduced under the suppression of TERT. Similar results were obtained with the suppression of either BRG1 (also called SMARCA4) or nucleostemin, which are proteins interacting with TERT and functioning beyond telomeres. These results suggest that TERT regulates microRNAs at the very early phases in their biogenesis, presumably through non-telomerase mechanism(s).
Figures

References
-
- Loffler D., Brocke-Heidrich K., Pfeifer G., Stocsits C., Hackermuller J., Kretzschmar A.K., Burger R., Gramatzki M., Blumert C., Bauer K., et al. Interleukin-6 dependent survival of multiple myeloma cells involves the Stat3-mediated induction of microRNA-21 through a highly conserved enhancer. Blood. 2007;110:1330–1333. doi: 10.1182/blood-2007-03-081133. - DOI - PubMed
-
- Brock M., Trenkmann M., Gay R.E., Michel B.A., Gay S., Fischler M., Ulrich S., Speich R., Huber L.C. Interleukin-6 modulates the expression of the bone morphogenic protein receptor type II through a novel STAT3-microRNA cluster 17/92 pathway. Circ. Res. 2009;104:1184–1191. doi: 10.1161/CIRCRESAHA.109.197491. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

