ASAR15, A cis-acting locus that controls chromosome-wide replication timing and stability of human chromosome 15
- PMID: 25569254
- PMCID: PMC4287527
- DOI: 10.1371/journal.pgen.1004923
ASAR15, A cis-acting locus that controls chromosome-wide replication timing and stability of human chromosome 15
Abstract
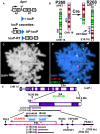
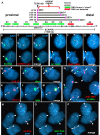
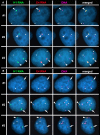
DNA replication initiates at multiple sites along each mammalian chromosome at different times during each S phase, following a temporal replication program. We have used a Cre/loxP-based strategy to identify cis-acting elements that control this replication-timing program on individual human chromosomes. In this report, we show that rearrangements at a complex locus at chromosome 15q24.3 result in delayed replication and structural instability of human chromosome 15. Characterization of this locus identified long, RNA transcripts that are retained in the nucleus and form a "cloud" on one homolog of chromosome 15. We also found that this locus displays asynchronous replication that is coordinated with other random monoallelic genes on chromosome 15. We have named this locus ASynchronous replication and Autosomal RNA on chromosome 15, or ASAR15. Previously, we found that disruption of the ASAR6 lincRNA gene results in delayed replication, delayed mitotic condensation and structural instability of human chromosome 6. Previous studies in the mouse found that deletion of the Xist gene, from the X chromosome in adult somatic cells, results in a delayed replication and instability phenotype that is indistinguishable from the phenotype caused by disruption of either ASAR6 or ASAR15. In addition, delayed replication and chromosome instability were detected following structural rearrangement of many different human or mouse chromosomes. These observations suggest that all mammalian chromosomes contain similar cis-acting loci. Thus, under this scenario, all mammalian chromosomes contain four distinct types of essential cis-acting elements: origins, telomeres, centromeres and "inactivation/stability centers", all functioning to promote proper replication, segregation and structural stability of each chromosome.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Nichols WW, A Levan, P Aula, E Norrby (1964) Extreme chromosome breakage induced by measles virus in different in vitro systems. Hereditas 51: 380.
-
- Nichols WW, A Levan, P Aula, E Norrby (1965) Chromosome damage associated with measles virus in vitro. Hereditas 54: 101. - PubMed
-
- Johnson RT, Rao PN (1970) Mammalian cell fusion: induction of premature chromosome condensation in interphase nuclei. Nature 226: 717–722. - PubMed
-
- Sperling K, Rao PN (1974) The phenomenon of premature chromosome condensation: its relevance to basic and applied research. Humangenetik 23: 235–258. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

