A new family of secreted toxins in pathogenic Neisseria species
- PMID: 25569427
- PMCID: PMC4287609
- DOI: 10.1371/journal.ppat.1004592
A new family of secreted toxins in pathogenic Neisseria species
Abstract
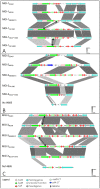
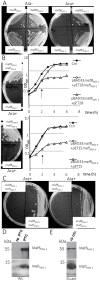
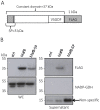
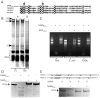
The genus Neisseria includes both commensal and pathogenic species which are genetically closely related. However, only meningococcus and gonococcus are important human pathogens. Very few toxins are known to be secreted by pathogenic Neisseria species. Recently, toxins secreted via type V secretion system and belonging to the widespread family of contact-dependent inhibition (CDI) toxins have been described in numerous species including meningococcus. In this study, we analyzed loci containing the maf genes in N. meningitidis and N. gonorrhoeae and proposed a novel uniform nomenclature for maf genomic islands (MGIs). We demonstrated that mafB genes encode secreted polymorphic toxins and that genes immediately downstream of mafB encode a specific immunity protein (MafI). We focused on a MafB toxin found in meningococcal strain NEM8013 and characterized its EndoU ribonuclease activity. maf genes represent 2% of the genome of pathogenic Neisseria, and are virtually absent from non-pathogenic species, thus arguing for an important biological role. Indeed, we showed that overexpression of one of the four MafB toxins of strain NEM8013 provides an advantage in competition assays, suggesting a role of maf loci in niche adaptation.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Comment in
-
Characterization of the Maf family of polymorphic toxins in pathogenic Neisseria species.Microb Cell. 2015 Mar 2;2(3):88-90. doi: 10.15698/mic2015.03.194. Microb Cell. 2015. PMID: 28357281 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

