Transposable elements contribute to activation of maize genes in response to abiotic stress
- PMID: 25569788
- PMCID: PMC4287451
- DOI: 10.1371/journal.pgen.1004915
Transposable elements contribute to activation of maize genes in response to abiotic stress
Erratum in
-
Correction: Transposable Elements Contribute to Activation of Maize Genes in Response to Abiotic Stress.PLoS Genet. 2015 Oct 9;11(10):e1005566. doi: 10.1371/journal.pgen.1005566. eCollection 2015 Oct. PLoS Genet. 2015. PMID: 26452261 Free PMC article. No abstract available.
Abstract
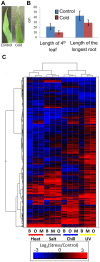
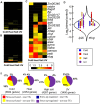
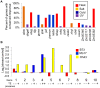
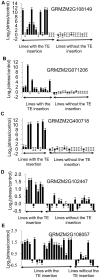
Transposable elements (TEs) account for a large portion of the genome in many eukaryotic species. Despite their reputation as "junk" DNA or genomic parasites deleterious for the host, TEs have complex interactions with host genes and the potential to contribute to regulatory variation in gene expression. It has been hypothesized that TEs and genes they insert near may be transcriptionally activated in response to stress conditions. The maize genome, with many different types of TEs interspersed with genes, provides an ideal system to study the genome-wide influence of TEs on gene regulation. To analyze the magnitude of the TE effect on gene expression response to environmental changes, we profiled gene and TE transcript levels in maize seedlings exposed to a number of abiotic stresses. Many genes exhibit up- or down-regulation in response to these stress conditions. The analysis of TE families inserted within upstream regions of up-regulated genes revealed that between four and nine different TE families are associated with up-regulated gene expression in each of these stress conditions, affecting up to 20% of the genes up-regulated in response to abiotic stress, and as many as 33% of genes that are only expressed in response to stress. Expression of many of these same TE families also responds to the same stress conditions. The analysis of the stress-induced transcripts and proximity of the transposon to the gene suggests that these TEs may provide local enhancer activities that stimulate stress-responsive gene expression. Our data on allelic variation for insertions of several of these TEs show strong correlation between the presence of TE insertions and stress-responsive up-regulation of gene expression. Our findings suggest that TEs provide an important source of allelic regulatory variation in gene response to abiotic stress in maize.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
-
- McClintock B (1956) Controlling Elements and the Gene. Cold Spring Harbor Symposia on Quantitative Biology 21: 197–216. - PubMed
-
- Wicker T, Sabot F, Hua-Van A, Bennetzen JL, Capy P, et al. (2007) A unified classification system for eukaryotic transposable elements. Nat Rev Genet 8: 973–982. - PubMed
-
- Feschotte C, Jiang N, Wessler S (2002) Plant transposable elements: where genetics meets genomics. Nat Rev Genet 3: 329–341. - PubMed
-
- McClintock B (1984) The significance of responses of the genome to challenge. Science 226: 792–801. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

