Identification of a human papillomavirus-associated oncogenic miRNA panel in human oropharyngeal squamous cell carcinoma validated by bioinformatics analysis of the Cancer Genome Atlas
- PMID: 25572154
- PMCID: PMC4348472
- DOI: 10.1016/j.ajpath.2014.11.018
Identification of a human papillomavirus-associated oncogenic miRNA panel in human oropharyngeal squamous cell carcinoma validated by bioinformatics analysis of the Cancer Genome Atlas
Abstract
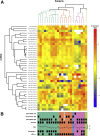
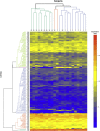
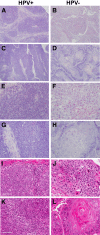
High-risk human papillomavirus (HPV) is a causative agent for an increasing subset of oropharyngeal squamous cell carcinomas (OPSCCs), and current evidence supports these tumors as having identifiable risk factors and improved response to therapy. However, the biochemical and molecular alterations underlying the pathobiology of HPV-associated OPSCC (designated HPV(+) OPSCC) remain unclear. Herein, we profile miRNA expression patterns in HPV(+) OPSCC to provide a more detailed understanding of pathologic molecular events and to identify biomarkers that may have applicability for early diagnosis, improved staging, and prognostic stratification. Differentially expressed miRNAs were identified in RNA isolated from an initial clinical cohort of HPV(+/-) OPSCC tumors by quantitative PCR-based miRNA profiling. This oncogenic miRNA panel was validated using miRNA sequencing and clinical data from The Cancer Genome Atlas and miRNA in situ hybridization. The HPV-associated oncogenic miRNA panel has potential utility in diagnosis and disease stratification and in mechanistic elucidation of molecular factors that contribute to OPSCC development, progression, and differential response to therapy.
Copyright © 2015 American Society for Investigative Pathology. Published by Elsevier Inc. All rights reserved.
Figures





References
-
- Gillison M.L., D'Souza G., Westra W., Sugar E., Xiao W., Begum S., Viscidi R. Distinct risk factor profiles for human papillomavirus type 16-positive and human papillomavirus type 16-negative head and neck cancers. J Natl Cancer Inst. 2008;100:407–420. - PubMed
-
- Ang K.K., Harris J., Wheeler R., Weber R., Rosenthal D.I., Nguyen-Tân P.F., Westra W.H., Chung C.H., Jordan R.C., Lu C., Kim H., Axelrod R., Silverman C.C., Redmond K.P., Gillison M.L. Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med. 2010;363:24–35. - PMC - PubMed
-
- Chaturvedi A.K., Engels E.A., Pfeiffer R.M., Hernandez B.Y., Xiao W., Kim E., Jiang B., Goodman M.T., Sibug-Saber M., Cozen W., Liu L., Lynch C.F., Wentzensen N., Jordan R.C., Altekruse S., Anderson W.F., Rosenberg P.S., Gillison M.L. Human papillomavirus and rising oropharyngeal cancer incidence in the United States. J Clin Oncol. 2011;29:4294–4301. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

