Phosphatidylinositol 4,5-bisphosphate clusters the cell adhesion molecule CD44 and assembles a specific CD44-Ezrin heterocomplex, as revealed by small angle neutron scattering
- PMID: 25572402
- PMCID: PMC4358296
- DOI: 10.1074/jbc.M114.589523
Phosphatidylinositol 4,5-bisphosphate clusters the cell adhesion molecule CD44 and assembles a specific CD44-Ezrin heterocomplex, as revealed by small angle neutron scattering
Abstract
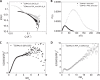
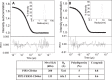
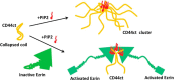
The cell adhesion molecule CD44 regulates diverse cellular functions, including cell-cell and cell-matrix interaction, cell motility, migration, differentiation, and growth. In cells, CD44 co-localizes with the membrane-cytoskeleton adapter protein Ezrin that links the CD44 assembled receptor signaling complexes to the cytoskeletal actin network, which organizes the spatial and temporal localization of signaling events. Here we report that the cytoplasmic tail of CD44 (CD44ct) is largely disordered. Upon binding to the signaling lipid phosphatidylinositol 4,5-bisphosphate (PIP2), CD44ct clusters into aggregates. Further, contrary to the generally accepted model, CD44ct does not bind directly to the FERM domain of Ezrin or to the full-length Ezrin but only forms a complex with FERM or with the full-length Ezrin in the presence of PIP2. Using contrast variation small angle neutron scattering, we show that PIP2 mediates the assembly of a specific heterotetramer complex of CD44ct with Ezrin. This study reveals the role of PIP2 in clustering CD44 and in assembling multimeric CD44-Ezrin complexes. We hypothesize that polyvalent electrostatic interactions are responsible for the assembly of CD44 clusters and the multimeric PIP2-CD44-Ezrin complexes.
Keywords: CD44; Cell Adhesion; Cell Adhesion Molecule; Ezrin; Neutron Scattering; Phosphatidylinositol.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures







References
-
- Martin T. A., Harrison G., Mansel R. E., Jiang W. G. (2003) The role of the CD44/Ezrin complex in cancer metastasis. Crit. Rev. Oncol. Hematol. 46, 165–186 - PubMed
-
- Ponta H., Sherman L., Herrlich P. A. (2003) CD44: From adhesion molecules to signalling regulators. Nat. Rev. Mol. Cell Biol. 4, 33–45 - PubMed
-
- Thorne R. F., Legg J. W., Isacke C. M. (2004) The role of the CD44 transmembrane and cytoplasmic domains in co-ordinating adhesive and signalling events. J. Cell Sci. 117, 373–380 - PubMed
-
- Toole B. P. (2004) Hyaluronan: from extracellular glue to pericellular cue. Nat. Rev. Cancer 4, 528–539 - PubMed
-
- Bourguignon L. Y., Zhu H., Shao L., Chen Y.-W. (2001) CD44 interaction with c-Src kinase promotes cortactin-mediated cytoskeleton function and hyaluronic acid-dependent ovarian tumor cell migration. J. Biol. Chem. 276, 7327–7336 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

