Identification of hub subnetwork based on topological features of genes in breast cancer
- PMID: 25573623
- PMCID: PMC4314413
- DOI: 10.3892/ijmm.2014.2057
Identification of hub subnetwork based on topological features of genes in breast cancer
Abstract
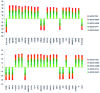
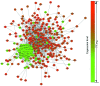
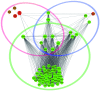
The aim of this study was to provide functional insight into the identification of hub subnetworks by aggregating the behavior of genes connected in a protein-protein interaction (PPI) network. We applied a protein network-based approach to identify subnetworks which may provide new insight into the functions of pathways involved in breast cancer rather than individual genes. Five groups of breast cancer data were downloaded and analyzed from the Gene Expression Omnibus (GEO) database of high-throughput gene expression data to identify gene signatures using the genome-wide global significance (GWGS) method. A PPI network was constructed using Cytoscape and clusters that focused on highly connected nodes were obtained using the molecular complex detection (MCODE) clustering algorithm. Pathway analysis was performed to assess the functional relevance of selected gene signatures based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. Topological centrality was used to characterize the biological importance of gene signatures, pathways and clusters. The results revealed that, cluster1, as well as the cell cycle and oocyte meiosis pathways were significant subnetworks in the analysis of degree and other centralities, in which hub nodes mostly distributed. The most important hub nodes, with top ranked centrality, were also similar with the common genes from the above three subnetwork intersections, which was viewed as a hub subnetwork with more reproducible than individual critical genes selected without network information. This hub subnetwork attributed to the same biological process which was essential in the function of cell growth and death. This increased the accuracy of identifying gene interactions that took place within the same functional process and was potentially useful for the development of biomarkers and networks for breast cancer.
Figures



Similar articles
-
Identifying hub genes and dysregulated pathways in hepatocellular carcinoma.Eur Rev Med Pharmacol Sci. 2015 Feb;19(4):592-601. Eur Rev Med Pharmacol Sci. 2015. PMID: 25753876
-
Incorporating topological information for predicting robust cancer subnetwork markers in human protein-protein interaction network.BMC Bioinformatics. 2016 Oct 6;17(Suppl 13):351. doi: 10.1186/s12859-016-1224-1. BMC Bioinformatics. 2016. PMID: 27766944 Free PMC article.
-
Identification of hub genes and pathways associated with retinoblastoma based on co-expression network analysis.Genet Mol Res. 2015 Dec 8;14(4):16151-61. doi: 10.4238/2015.December.8.4. Genet Mol Res. 2015. PMID: 26662407
-
IODNE: An integrated optimization method for identifying the deregulated subnetwork for precision medicine in cancer.CPT Pharmacometrics Syst Pharmacol. 2017 Mar;6(3):168-176. doi: 10.1002/psp4.12167. Epub 2017 Mar 7. CPT Pharmacometrics Syst Pharmacol. 2017. PMID: 28266149 Free PMC article. Review.
-
Gene Prioritization and Network Topology Analysis of Targeted Genes for Acquired Taxane Resistance by Meta-Analysis.Crit Rev Eukaryot Gene Expr. 2019;29(6):581-597. doi: 10.1615/CritRevEukaryotGeneExpr.2019026317. Crit Rev Eukaryot Gene Expr. 2019. PMID: 32422012 Review.
Cited by
-
Prediction of new candidate proteins and analysis of sub-modules and protein hubs associated with seed development in rice (Oryza sativa) using an ensemble network-based systems biology approach.BMC Plant Biol. 2025 May 8;25(1):604. doi: 10.1186/s12870-025-06595-7. BMC Plant Biol. 2025. PMID: 40340735 Free PMC article.
-
Repurposing Nirmatrelvir for Hepatocellular Carcinoma: Network Pharmacology and Molecular Dynamics Simulations Identify HDAC3 as a Key Molecular Target.Pharmaceuticals (Basel). 2025 Jul 31;18(8):1144. doi: 10.3390/ph18081144. Pharmaceuticals (Basel). 2025. PMID: 40872535 Free PMC article.
-
Meta-analysis of transcriptomics data identifies potential biomarkers and their associated regulatory networks in gallbladder cancer.Gastroenterol Hepatol Bed Bench. 2022;15(4):311-325. doi: 10.22037/ghfbb.v15i4.2292. Gastroenterol Hepatol Bed Bench. 2022. PMID: 36762219 Free PMC article. Review.
-
Genome-scale analysis identifies SERPINE1 and SPARC as diagnostic and prognostic biomarkers in gastric cancer.Onco Targets Ther. 2018 Oct 15;11:6969-6980. doi: 10.2147/OTT.S173934. eCollection 2018. Onco Targets Ther. 2018. PMID: 30410354 Free PMC article.
-
Screening of the prognostic targets for breast cancer based co-expression modules analysis.Mol Med Rep. 2017 Oct;16(4):4038-4044. doi: 10.3892/mmr.2017.7063. Epub 2017 Jul 21. Mol Med Rep. 2017. PMID: 28731166 Free PMC article.
References
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

