Characterization of the host response to pichinde virus infection in the Syrian golden hamster by species-specific kinome analysis
- PMID: 25573744
- PMCID: PMC4349984
- DOI: 10.1074/mcp.M114.045443
Characterization of the host response to pichinde virus infection in the Syrian golden hamster by species-specific kinome analysis
Abstract
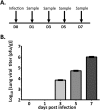
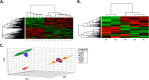
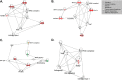
The Syrian golden hamster has been increasingly used to study viral hemorrhagic fever (VHF) pathogenesis and countermeasure efficacy. As VHFs are a global health concern, well-characterized animal models are essential for both the development of therapeutics and vaccines as well as for increasing our understanding of the molecular events that underlie viral pathogenesis. However, the paucity of reagents or platforms that are available for studying hamsters at a molecular level limits the ability to extract biological information from this important animal model. As such, there is a need to develop platforms/technologies for characterizing host responses of hamsters at a molecular level. To this end, we developed hamster-specific kinome peptide arrays to characterize the molecular host response of the Syrian golden hamster. After validating the functionality of the arrays using immune agonists of defined signaling mechanisms (lipopolysaccharide (LPS) and tumor necrosis factor (TNF)-α), we characterized the host response in a hamster model of VHF based on Pichinde virus (PICV(1)) infection by performing temporal kinome analysis of lung tissue. Our analysis revealed key roles for vascular endothelial growth factor (VEGF), interleukin (IL) responses, nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) signaling, and Toll-like receptor (TLR) signaling in the response to PICV infection. These findings were validated through phosphorylation-specific Western blot analysis. Overall, we have demonstrated that hamster-specific kinome arrays are a robust tool for characterizing the species-specific molecular host response in a VHF model. Further, our results provide key insights into the hamster host response to PICV infection and will inform future studies with high-consequence VHF pathogens.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures


Similar articles
-
Development of infectious clones for virulent and avirulent pichinde viruses: a model virus to study arenavirus-induced hemorrhagic fevers.J Virol. 2009 Jul;83(13):6357-62. doi: 10.1128/JVI.00019-09. Epub 2009 Apr 22. J Virol. 2009. PMID: 19386714 Free PMC article.
-
The nucleoprotein of Pichinde virus expressed by a vaccinia-Pichinde virus recombinant partially protects hamsters from lethal virus challenge.Arch Virol. 1994;139(1-2):23-36. doi: 10.1007/BF01309452. Arch Virol. 1994. PMID: 7826212
-
Pirital virus (Arenaviridae) infection in the syrian golden hamster, Mesocricetus auratus: a new animal model for arenaviral hemorrhagic fever.Am J Trop Med Hyg. 2001 Mar-Apr;64(3-4):111-8. doi: 10.4269/ajtmh.2001.64.111. Am J Trop Med Hyg. 2001. PMID: 11442204
-
Systems kinomics for characterizing host responses to high-consequence pathogens at the NIH/NIAID Integrated Research Facility-Frederick.Pathog Dis. 2014 Jul;71(2):190-98. doi: 10.1111/2049-632X.12163. Epub 2014 Apr 10. Pathog Dis. 2014. PMID: 24585711 Free PMC article. Review.
-
From Beef to Bees: High-Throughput Kinome Analysis to Understand Host Responses of Livestock Species to Infectious Diseases and Industry-Associated Stress.Front Immunol. 2020 May 15;11:765. doi: 10.3389/fimmu.2020.00765. eCollection 2020. Front Immunol. 2020. PMID: 32499776 Free PMC article. Review.
Cited by
-
Rodent-Adapted Filoviruses and the Molecular Basis of Pathogenesis.J Mol Biol. 2016 Aug 28;428(17):3449-66. doi: 10.1016/j.jmb.2016.05.008. Epub 2016 May 14. J Mol Biol. 2016. PMID: 27189922 Free PMC article. Review.
-
TRIM2, a novel member of the antiviral family, limits New World arenavirus entry.PLoS Biol. 2019 Feb 6;17(2):e3000137. doi: 10.1371/journal.pbio.3000137. eCollection 2019 Feb. PLoS Biol. 2019. PMID: 30726215 Free PMC article.
-
Integration of Global Analyses of Host Molecular Responses with Clinical Data To Evaluate Pathogenesis and Advance Therapies for Emerging and Re-emerging Viral Infections.ACS Infect Dis. 2016 Nov 11;2(11):787-799. doi: 10.1021/acsinfecdis.6b00104. Epub 2016 Aug 5. ACS Infect Dis. 2016. PMID: 27933782 Free PMC article. Review.
-
Syrian Hamster as an Animal Model for the Study on Infectious Diseases.Front Immunol. 2019 Oct 1;10:2329. doi: 10.3389/fimmu.2019.02329. eCollection 2019. Front Immunol. 2019. PMID: 31632404 Free PMC article. Review.
-
Methods and approaches to disease mechanisms using systems kinomics.Synth Syst Biotechnol. 2017 Dec 18;3(1):34-43. doi: 10.1016/j.synbio.2017.12.004. eCollection 2018 Mar. Synth Syst Biotechnol. 2017. PMID: 29911197 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

