MATR3 disruption in human and mouse associated with bicuspid aortic valve, aortic coarctation and patent ductus arteriosus
- PMID: 25574029
- PMCID: PMC4380077
- DOI: 10.1093/hmg/ddv004
MATR3 disruption in human and mouse associated with bicuspid aortic valve, aortic coarctation and patent ductus arteriosus
Abstract
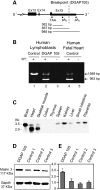
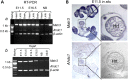
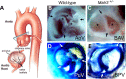
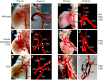
Cardiac left ventricular outflow tract (LVOT) defects represent a common but heterogeneous subset of congenital heart disease for which gene identification has been difficult. We describe a 46,XY,t(1;5)(p36.11;q31.2)dn translocation carrier with pervasive developmental delay who also exhibited LVOT defects, including bicuspid aortic valve (BAV), coarctation of the aorta (CoA) and patent ductus arteriosus (PDA). The 1p breakpoint disrupts the 5' UTR of AHDC1, which encodes AT-hook DNA-binding motif containing-1 protein, and AHDC1-truncating mutations have recently been described in a syndrome that includes developmental delay, but not congenital heart disease [Xia, F., Bainbridge, M.N., Tan, T.Y., Wangler, M.F., Scheuerle, A.E., Zackai, E.H., Harr, M.H., Sutton, V.R., Nalam, R.L., Zhu, W. et al. (2014) De Novo truncating mutations in AHDC1 in individuals with syndromic expressive language delay, hypotonia, and sleep apnea. Am. J. Hum. Genet., 94, 784-789]. On the other hand, the 5q translocation breakpoint disrupts the 3' UTR of MATR3, which encodes the nuclear matrix protein Matrin 3, and mouse Matr3 is strongly expressed in neural crest, developing heart and great vessels, whereas Ahdc1 is not. To further establish MATR3 3' UTR disruption as the cause of the proband's LVOT defects, we prepared a mouse Matr3(Gt-ex13) gene trap allele that disrupted the 3' portion of the gene. Matr3(Gt-ex13) homozygotes are early embryo lethal, but Matr3(Gt-ex13) heterozygotes exhibit incompletely penetrant BAV, CoA and PDA phenotypes similar to those in the human proband, as well as ventricular septal defect (VSD) and double-outlet right ventricle (DORV). Both the human MATR3 translocation breakpoint and the mouse Matr3(Gt-ex13) gene trap insertion disturb the polyadenylation of MATR3 transcripts and alter Matrin 3 protein expression, quantitatively or qualitatively. Thus, subtle perturbations in Matrin 3 expression appear to cause similar LVOT defects in human and mouse.
© The Author 2015. Published by Oxford University Press.
Figures





References
-
- Jenkins K.J., Correa A., Feinstein J.A., Botto L., Britt A.E., Daniels S.R., Elixson M., Warnes C.A., Webb C.L. (2007) Noninherited risk factors and congenital cardiovascular defects: current knowledge: a scientific statement from the American Heart Association Council on Cardiovascular Disease in the Young: endorsed by the American Academy of Pediatrics. Circulation, 115, 2995–3014. - PubMed
-
- Pierpont M.E., Basson C.T., Benson D.W., Jr, Gelb B.D., Giglia T.M., Goldmuntz E., McGee G., Sable C.A., Srivastava D., Webb C.L. (2007) Genetic basis for congenital heart defects: current knowledge: a scientific statement from the American Heart Association Congenital Cardiac Defects Committee, Council on Cardiovascular Disease in the Young: endorsed by the American Academy of Pediatrics. Circulation, 115, 3015–3028. - PubMed
-
- Siu S.C., Silversides C.K. (2010) Bicuspid aortic valve disease. J. Am. Coll. Cardiol., 55, 2789–2800. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

