Diverse protist grazers select for virulence-related traits in Legionella
- PMID: 25575308
- PMCID: PMC4478701
- DOI: 10.1038/ismej.2014.248
Diverse protist grazers select for virulence-related traits in Legionella
Abstract
It is generally accepted that selection for resistance to grazing by protists has contributed to the evolution of Legionella pneumophila as a pathogen. Grazing resistance is becoming more generally recognized as having an important role in the ecology and evolution of bacterial pathogenesis. However, selection for grazing resistance presupposes the existence of protist grazers that provide the selective pressure. To determine whether there are protists that graze on pathogenic Legionella species, we investigated the existence of such organisms in a variety of environmental samples. We isolated and characterized diverse protists that graze on L. pneumophila and determined the effects of adding L. pneumophila on the protist community structures in microcosms made from these environmental samples. Several unrelated organisms were able to graze efficiently on L. pneumophila. The community structures of all samples were markedly altered by the addition of L. pneumophila. Surprisingly, some of the Legionella grazers were closely related to species that are known hosts for L. pneumophila, indicating the presence of unknown specificity determinants for this interaction. These results provide the first direct support for the hypothesis that protist grazers exert selective pressure on Legionella to acquire and retain adaptations that contribute to survival, and that these properties are relevant to the ability of the bacteria to cause disease in people. We also report a novel mechanism of killing of amoebae by one Legionella species that requires an intact Type IV secretion system but does not involve intracellular replication. We refer to this phenomenon as 'food poisoning'.
Figures

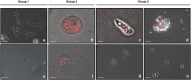
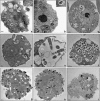

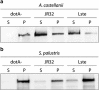
References
-
- Adl SM, Simpson AG, Farmer MA, Andersen RA, Anderson OR, Barta JR, et al. The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J Eukaryot Microbiol. 2005;52:399–451. - PubMed
-
- Anderson OR, Wang W, Faucher SP, Bi K, Shuman HA. A new Heterolobosean amoeba Solumitrus palustris n. g., n. sp. isolated from freshwater marsh soil. J Eukaryot Microbiol. 2011;58:60–67. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

