Importin-α-mediated nucleolar localization of potato mop-top virus TRIPLE GENE BLOCK1 (TGB1) protein facilitates virus systemic movement, whereas TGB1 self-interaction is required for cell-to-cell movement in Nicotiana benthamiana
- PMID: 25576325
- PMCID: PMC4348779
- DOI: 10.1104/pp.114.254938
Importin-α-mediated nucleolar localization of potato mop-top virus TRIPLE GENE BLOCK1 (TGB1) protein facilitates virus systemic movement, whereas TGB1 self-interaction is required for cell-to-cell movement in Nicotiana benthamiana
Abstract
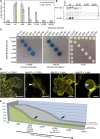
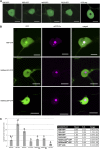
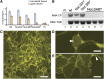
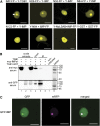
Recently, it has become evident that nucleolar passage of movement proteins occurs commonly in a number of plant RNA viruses that replicate in the cytoplasm. Systemic movement of Potato mop-top virus (PMTV) involves two viral transport forms represented by a complex of viral RNA and TRIPLE GENE BLOCK1 (TGB1) movement protein and by polar virions that contain the minor coat protein and TGB1 attached to one extremity. The integrity of polar virions ensures the efficient movement of RNA-CP, which encodes the virus coat protein. Here, we report the involvement of nuclear transport receptors belonging to the importin-α family in nucleolar accumulation of the PMTV TGB1 protein and, subsequently, in the systemic movement of the virus. Virus-induced gene silencing of two importin-α paralogs in Nicotiana benthamiana resulted in significant reduction of TGB1 accumulation in the nucleus, decreasing the accumulation of the virus progeny in upper leaves and the loss of systemic movement of RNA-CP. PMTV TGB1 interacted with importin-α in N. benthamiana, which was detected by bimolecular fluorescence complementation in the nucleoplasm and nucleolus. The interaction was mediated by two nucleolar localization signals identified by bioinformatics and mutagenesis in the TGB1 amino-terminal domain. Our results showed that while TGB1 self-interaction is needed for cell-to-cell movement, importin-α-mediated nucleolar targeting of TGB1 is an essential step in establishing the efficient systemic infection of the entire plant. These results enabled the identification of two separate domains in TGB1: an internal domain required for TGB1 self-interaction and cell-to-cell movement and the amino-terminal domain required for importin-α interaction in plants, nucleolar targeting, and long-distance movement.
© 2015 American Society of Plant Biologists. All Rights Reserved.
Figures

References
-
- Arif M, Torrance L, Reavy B (1995) Acquisition and transmission of potato mop-top furovirus by a culture of Spongospora subterranea f.sp subterranea derived from a single cystosorus. Ann Appl Biol 126: 493–503
-
- Barneche F, Steinmetz F, Echeverría M (2000) Fibrillarin genes encode both a conserved nucleolar protein and a novel small nucleolar RNA involved in ribosomal RNA methylation in Arabidopsis thaliana. J Biol Chem 275: 27212–27220 - PubMed
-
- Chiba S, Hleibieh K, Delbianco A, Klein E, Ratti C, Ziegler-Graff V, Bouzoubaa S, Gilmer D (2013) The benyvirus RNA silencing suppressor is essential for long-distance movement, requires both zinc-finger and NoLS basic residues but not a nucleolar localization for its silencing-suppression activity. Mol Plant Microbe Interact 26: 168–181 - PubMed
-
- Dosztányi Z, Csizmok V, Tompa P, Simon I (2005) IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics 21: 3433–3434 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

