cis-Acting RNA elements in the hepatitis C virus RNA genome
- PMID: 25576644
- PMCID: PMC5553863
- DOI: 10.1016/j.virusres.2014.12.029
cis-Acting RNA elements in the hepatitis C virus RNA genome
Abstract
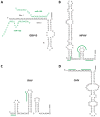
Hepatitis C virus (HCV) infection is a rapidly increasing global health problem with an estimated 170 million people infected worldwide. HCV is a hepatotropic, positive-sense RNA virus of the family Flaviviridae. As a positive-sense RNA virus, the HCV genome itself must serve as a template for translation, replication and packaging. The viral RNA must therefore be a dynamic structure that is able to readily accommodate structural changes to expose different regions of the genome to viral and cellular proteins to carry out the HCV life cycle. The ∼ 9600 nucleotide viral genome contains a single long open reading frame flanked by 5' and 3' non-coding regions that contain cis-acting RNA elements important for viral translation, replication and stability. Additional cis-acting RNA elements have also been identified in the coding sequences as well as in the 3' end of the negative-strand replicative intermediate. Herein, we provide an overview of the importance of these cis-acting RNA elements in the HCV life cycle.
Keywords: Hepatitis C virus; Internal ribosome entry site; cis-Acting RNA element; miR-122.
Copyright © 2015 Elsevier B.V. All rights reserved.
Figures

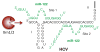
References
-
- Hoofnagle JH. Course and outcome of hepatitis C. Hepatology. 2002;36(5 Suppl 1):S21–9. - PubMed
-
- Lavanchy D. Evolving epidemiology of hepatitis C virus. Clinical microbiology and infection : the official publication of the European Society of Clinical Microbiology and Infectious Diseases. 2011;17(2):107–15. - PubMed
-
- Casey LC, Lee WM. Hepatitis C virus therapy update 2013. Curr Opin Gastroenterol. 2013;29(3):243–9. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

