Sexual differences in the sialomes of the zebra tick, Rhipicephalus pulchellus
- PMID: 25576852
- PMCID: PMC4374903
- DOI: 10.1016/j.jprot.2014.12.014
Sexual differences in the sialomes of the zebra tick, Rhipicephalus pulchellus
Abstract
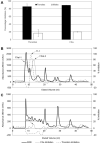
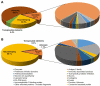
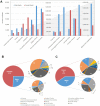
Ticks rely exclusively on vertebrate blood for their survival. During feeding ticks inject into their hosts a sophisticated salivary potion that overcomes host hemostasis and adverse inflammatory responses. These mediators may also enhance pathogen transmission. Knowledge of the tick salivary protein repertoire may lead to vaccine targets to disrupt feeding and/or parasite transmission as well as to the discovery of novel pharmacological agents. Male saliva may also assist reproduction because males use their mouthparts to lubricate and introduce their spermatophores into the females' genital pore. The analyses of the sialomes of male and female ticks independently allow us to understand the strategy used by each gender to feed successfully. We sequenced cDNA libraries from pools of salivary glands from adult male and female Rhipicephalus pulchellus feeding at different time points, using the Illumina HiSeq protocol. De novo assembly of a total of 241,229,128 paired-end reads lead to extraction of 50,460 coding sequences (CDS), 11,277 of which had more than 75% coverage to known transcripts, or represented novel sequences, and were submitted to GenBank. Additionally, we generated the proteome, from the salivary gland extracts of male and female R. pulchellus, yielding a total of 454 and 2063 proteins respectively which were identified by one or more peptides with at least 95% confidence. The data set is presented as an annotated hyperlinked Excel spreadsheet, describing 121 putative secreted protein families. Female and male specific transcripts were identified.
Biological significance: This annotated R. pulchellus database represents a mining field for future experiments involving the resolution of time-dependent transcript expression in this tick species, as well as to define novel vaccine targets and discover novel pharmaceuticals. Gender specific proteins may represent different repertoires of pharmacological reagents to assist feeding by each sex, and in males may represent proteins that assist reproduction similarly to seminal proteins in other animals.
Keywords: Salivary glands; Tick; Transcriptome.
Copyright © 2015 Elsevier B.V. All rights reserved.
Figures




Similar articles
-
Gene Expression in the Salivary Gland of Rhipicephalus (Boophilus) microplus Fed on Tick-Susceptible and Tick-Resistant Hosts.Front Cell Infect Microbiol. 2020 Jan 21;9:477. doi: 10.3389/fcimb.2019.00477. eCollection 2019. Front Cell Infect Microbiol. 2020. PMID: 32039052 Free PMC article.
-
RNA-seq analysis and gene expression dynamics in the salivary glands of the argasid tick Ornithodoros erraticus along the trophogonic cycle.Parasit Vectors. 2021 Mar 20;14(1):170. doi: 10.1186/s13071-021-04671-z. Parasit Vectors. 2021. PMID: 33743776 Free PMC article.
-
An Insight into the Sialome of the Lone Star Tick, Amblyomma americanum, with a Glimpse on Its Time Dependent Gene Expression.PLoS One. 2015 Jul 1;10(7):e0131292. doi: 10.1371/journal.pone.0131292. eCollection 2015. PLoS One. 2015. PMID: 26131772 Free PMC article.
-
The Essential Role of Tick Salivary Glands and Saliva in Tick Feeding and Pathogen Transmission.Front Cell Infect Microbiol. 2017 Jun 22;7:281. doi: 10.3389/fcimb.2017.00281. eCollection 2017. Front Cell Infect Microbiol. 2017. PMID: 28690983 Free PMC article. Review.
-
Tick salivary compounds: their role in modulation of host defences and pathogen transmission.Front Cell Infect Microbiol. 2013 Aug 20;3:43. doi: 10.3389/fcimb.2013.00043. eCollection 2013. Front Cell Infect Microbiol. 2013. PMID: 23971008 Free PMC article. Review.
Cited by
-
Deciphering Biological Processes at the Tick-Host Interface Opens New Strategies for Treatment of Human Diseases.Front Physiol. 2019 Jul 3;10:830. doi: 10.3389/fphys.2019.00830. eCollection 2019. Front Physiol. 2019. PMID: 31333488 Free PMC article. Review.
-
Partial characterization of a novel anti-inflammatory protein from salivary gland extract of Hyalomma anatolicum anatolicum 77Acari: Ixodidae) ticks.Vet World. 2015 Jun;8(6):772-6. doi: 10.14202/vetworld.2015.772-776. Epub 2015 Jun 24. Vet World. 2015. PMID: 27065646 Free PMC article.
-
Sialomes and Mialomes: A Systems-Biology View of Tick Tissues and Tick-Host Interactions.Trends Parasitol. 2016 Mar;32(3):242-254. doi: 10.1016/j.pt.2015.10.002. Epub 2015 Oct 28. Trends Parasitol. 2016. PMID: 26520005 Free PMC article. Review.
-
Sialotranscriptomics of the argasid tick Ornithodoros moubata along the trophogonic cycle.PLoS Negl Trop Dis. 2021 Feb 5;15(2):e0009105. doi: 10.1371/journal.pntd.0009105. eCollection 2021 Feb. PLoS Negl Trop Dis. 2021. PMID: 33544727 Free PMC article.
-
Coxiella Endosymbiont of Rhipicephalus microplus Modulates Tick Physiology With a Major Impact in Blood Feeding Capacity.Front Microbiol. 2022 May 3;13:868575. doi: 10.3389/fmicb.2022.868575. eCollection 2022. Front Microbiol. 2022. PMID: 35591999 Free PMC article.
References
-
- Nava S, Guglielmone AA, Mangold AJ. An overview of systematics and evolution of ticks. Front Biosci. 2009;14:2857–77. - PubMed
-
- Mans BJ, Francischetti IMB. Sialomic perspectives on the evolution of blood-feeding behavior in Arthropods: Future therapeutics by natural design. In: Kini RM, Clemetson KJ, Markland FS, McLane MA, Morita T, editors. Toxins and Hemostasis: From Bench to Bed-side. Springer; 2010.
-
- Mans BJ, Neitz AW. Adaptation of ticks to a blood-feeding environment: evolution from a functional perspective. Insect Biochem Mol Biol. 2004;34:1–17. - PubMed
-
- Lehane MJ. The biology of blood-sucking in insects. 2 ed. Cambridge University Press; Cambridge: 2005.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

