Understanding host-adherent-invasive Escherichia coli interaction in Crohn's disease: opening up new therapeutic strategies
- PMID: 25580435
- PMCID: PMC4279263
- DOI: 10.1155/2014/567929
Understanding host-adherent-invasive Escherichia coli interaction in Crohn's disease: opening up new therapeutic strategies
Abstract
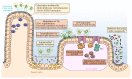
A trillion of microorganisms colonize the mammalian intestine. Most of them have coevolved with the host in a symbiotic relationship and some of them have developed strategies to promote their replication in the presence of competing microbiota. Recent evidence suggests that perturbation of the microbial community favors the emergence of opportunistic pathogens, in particular adherent-invasive Escherichia coli (AIEC) that can increase incidence and severity of gut inflammation in the context of Crohn's disease (CD). This review will report the importance of AIEC as triggers of intestinal inflammation, focusing on their impact on epithelial barrier function and stimulation of mucosal inflammation. Beyond manipulation of immune response, restoration of gut microbiota as a new treatment option for CD patients will be discussed.
Figures
References
-
- Qin J., Li R., Raes J., Arumugam M., Burgdorf K. S., Manichanh C., Nielsen T., Pons N., Levenez F., Yamada T., Mende D. R., Li J., Xu J., Li S., Li D., Cao J., Wang B., Liang H., Zheng H., Xie Y., Tap J., Lepage P., Bertalan M., Batto J.-M., Hansen T., Le Paslier D., Linneberg A., Nielsen H. B., Pelletier E., Renault P., Sicheritz-Ponten T., Turner K., Zhu H., Yu C., Li S., Jian M., Zhou Y., Li Y., Zhang X., Li S., Qin N., Yang H., Wang J., Brunak S., Doré J., Guarner F., Kristiansen K., Pedersen O., Parkhill J., Weissenbach J., Bork P., Ehrlich S. D., Wang J., Antolin M., Artiguenave F., Blottiere H., Borruel N., Bruls T., Casellas F., Chervaux C., Cultrone A., Delorme C., Denariaz G., Dervyn R., Forte M., Friss C., Van De Guchte M., Guedon E., Haimet F., Jamet A., Juste C., Kaci G., Kleerebezem M., Knol J., Kristensen M., Layec S., Le Roux K., Leclerc M., Maguin E., Melo Minardi R., Oozeer R., Rescigno M., Sanchez N., Tims S., Torrejon T., Varela E., De Vos W., Winogradsky Y., Zoetendal E. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464(7285):59–65. doi: 10.1038/nature08821. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

