Tetrameric Ctp1 coordinates DNA binding and DNA bridging in DNA double-strand-break repair
- PMID: 25580577
- PMCID: PMC4318798
- DOI: 10.1038/nsmb.2945
Tetrameric Ctp1 coordinates DNA binding and DNA bridging in DNA double-strand-break repair
Abstract
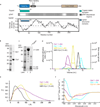
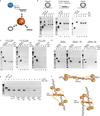
Ctp1 (also known as CtIP or Sae2) collaborates with Mre11-Rad50-Nbs1 to initiate repair of DNA double-strand breaks (DSBs), but its functions remain enigmatic. We report that tetrameric Schizosaccharomyces pombe Ctp1 contains multivalent DNA-binding and DNA-bridging activities. Through structural and biophysical analyses of the Ctp1 tetramer, we define the salient features of Ctp1 architecture: an N-terminal interlocking tetrameric helical dimer-of-dimers (THDD) domain and a central intrinsically disordered region (IDR) linked to C-terminal 'RHR' DNA-interaction motifs. The THDD, IDR and RHR are required for Ctp1 DNA-bridging activity in vitro, and both the THDD and RHR are required for efficient DSB repair in S. pombe. Our results establish non-nucleolytic roles of Ctp1 in binding and coordination of DSB-repair intermediates and suggest that ablation of human CtIP DNA binding by truncating mutations underlie the CtIP-linked Seckel and Jawad syndromes.
Figures





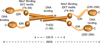
References
-
- Williams RS, Williams JS, Tainer JA. Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochem. Cell Biol. 2007;85:509–520. - PubMed
Online Methods References
-
- Stols L, et al. A new vector for high-throughput, ligation-independent cloning encoding a tobacco etch virus protease cleavage site. Protein Expr. Purif. 2002;25:8–15. - PubMed
-
- Sreerama N, Woody RW. A self-consistent method for the analysis of protein secondary structure from circular dichroism. Anal. Biochem. 1993;209:32–44. - PubMed
-
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in osciallation mode. In: Carter CW Jr, Sweet RM, editors. Methods in Enzymology. Vol. 276. Academic Press; 1997. pp. 307–326. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

