Innate immune pattern recognition: a cell biological perspective
- PMID: 25581309
- PMCID: PMC5146691
- DOI: 10.1146/annurev-immunol-032414-112240
Innate immune pattern recognition: a cell biological perspective
Abstract
Receptors of the innate immune system detect conserved determinants of microbial and viral origin. Activation of these receptors initiates signaling events that culminate in an effective immune response. Recently, the view that innate immune signaling events rely on and operate within a complex cellular infrastructure has become an important framework for understanding the regulation of innate immunity. Compartmentalization within this infrastructure provides the cell with the ability to assign spatial information to microbial detection and regulate immune responses. Several cell biological processes play a role in the regulation of innate signaling responses; at the same time, innate signaling can engage cellular processes as a form of defense or to promote immunological memory. In this review, we highlight these aspects of cell biology in pattern-recognition receptor signaling by focusing on signals that originate from the cell surface, from endosomal compartments, and from within the cytosol.
Keywords: ALR; CLR; NLR; RLR; TLR.
Figures
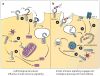
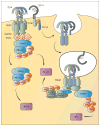
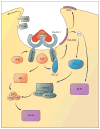
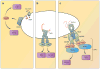

References
-
- Kumar H, Kawai T, Akira S. Pathogen recognition by the innate immune system. Int Rev Immunol. 2011;30(1):16–34. - PubMed
-
- Janeway CA. Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb Symp Quant Biol. 1989;54(Pt. 1):1–13. - PubMed
-
- Medzhitov R. Approaching the asymptote: 20 years later. Immunity. 2009;30(6):766–75. - PubMed
-
- Zelensky AN, Gready JE. The C-type lectin-like domain superfamily. FEBS J. 2005;272(24):6179–217. - PubMed
-
- Drummond RA, Brown GD. The role of Dectin-1 in the host defence against fungal infections. Curr Opin Microbiol. 2011;14(4):392–99. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

