Ammonia-oxidizing Bacteria of the Nitrosospira cluster 1 dominate over ammonia-oxidizing Archaea in oligotrophic surface sediments near the South Atlantic Gyre
- PMID: 25581373
- PMCID: PMC5008181
- DOI: 10.1111/1758-2229.12264
Ammonia-oxidizing Bacteria of the Nitrosospira cluster 1 dominate over ammonia-oxidizing Archaea in oligotrophic surface sediments near the South Atlantic Gyre
Abstract
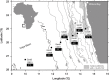
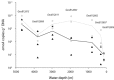
Sediments across the Namibian continental margin feature a strong microbial activity gradient at their surface. This is reflected in ammonium concentrations of < 10 μM in oligotrophic abyssal plain sediments near the South Atlantic Gyre compared with ammonium concentrations of > 700 μM in upwelling areas near the coast. Here we address changes in apparent abundance and structure of ammonia-oxidizing archaeal and bacterial communities (AOA and AOB) along a transect of seven sediment stations across the Namibian shelf by analysing their respective ammonia monooxygenase genes (amoA). The relative abundance of archaeal and bacterial amoA (g(-1) DNA) decreased with increasing ammonium concentrations, and bacterial amoA frequently outnumbered archaeal amoA at the sediment-water interface [0-1 cm below seafloor (cmbsf)]. In contrast, AOA were apparently as abundant as AOB or dominated in several deeper (> 10 cmbsf), anoxic sediment layers. Phylogenetic analyses showed a change within the AOA community along the transect, from two clusters without cultured representatives at the gyre to Nitrososphaera and Nitrosopumilus clusters in the upwelling region. AOB almost exclusively belonged to the Nitrosospira cluster 1. Our results suggest that this predominantly marine AOB lineage without cultured representatives can thrive at low ammonium concentrations and is active in the marine nitrogen cycle.
© 2015 The Authors. Environmental Microbiology Reports published by Society for Applied Microbiology and John Wiley & Sons Ltd.
Figures

References
-
- Berg, P. , Risgaard‐Petersen, N. , and Rysgaard, S. (1998) Interpretation of measured concentration profiles in sediment pore water. Limnol Oceanogr 43: 1500–1510.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

