BRCA1 and CtIP promote alternative non-homologous end-joining at uncapped telomeres
- PMID: 25582120
- PMCID: PMC4339125
- DOI: 10.15252/embj.201488947
BRCA1 and CtIP promote alternative non-homologous end-joining at uncapped telomeres
Erratum in
-
BRCA1 and CtIP promote alternative non-homologous end-joining at uncapped telomeres.EMBO J. 2015 Mar 12;34(6):828. doi: 10.15252/embj.201570610. EMBO J. 2015. PMID: 25766694 Free PMC article. No abstract available.
Abstract
Loss of telomere protection occurs during physiological cell senescence and ageing, due to attrition of telomeric repeats and insufficient retention of the telomere-binding factor TRF2. Subsequently formed telomere fusions trigger rampant genomic instability leading to cell death or tumorigenesis. Mechanistically, telomere fusions require either the classical non-homologous end-joining (C-NHEJ) pathway dependent on Ku70/80 and LIG4, or the alternative non-homologous end-joining (A-NHEJ), which relies on PARP1 and LIG3. Here, we show that the tumour suppressor BRCA1, together with its interacting partner CtIP, both acting in end resection, also promotes end-joining of uncapped telomeres. BRCA1 and CtIP do not function in the ATM-dependent telomere damage signalling, nor in telomere overhang removal, which are critical for telomere fusions by C-NHEJ. Instead, BRCA1 and CtIP act in the same pathway as LIG3 to promote joining of de-protected telomeres by A-NHEJ. Our work therefore ascribes novel roles for BRCA1 and CtIP in end-processing and fusion reactions at uncapped telomeres, underlining the complexity of DNA repair pathways that act at chromosome ends lacking protective structures. Moreover, A-NHEJ provides a mechanism of previously unanticipated significance in telomere dysfunction-induced genome instability.
Keywords: BRCA1/CtIP; TRF2; alternative non‐homologous end‐joining; telomere.
© 2015 The Authors. Published under the terms of the CC BY 4.0 license.
Figures

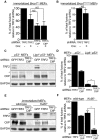
Immortalized Brca1F/− MEFs were infected with retroviruses expressing the indicated shRNAs and/or Cre recombinase, followed by selection with puromycin for 72 h. Cell extracts were prepared 48 h later and analysed by Western blotting as indicated. SMC1 and tubulin were used as loading controls. *non-specific band.
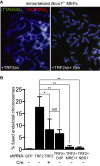
Cells treated as in (A) were fixed 48 h after selection and stained with an anti-53BP1 antibody (green). Telomeres were visualized with a Cy3-conjugated (CCCTAA)3-PNA probe (red). Yellow arrowheads point to 53BP1 foci that co-localize with telomeres.
Quantification of TIFs in cells treated as in (B). A minimum of 200 nuclei were scored for each sample. Error bars represent SD of two independent experiments. P-values were calculated using an unpaired two-tailed t-test. *P ≤ 0.05; NS, P > 0.05.
Immortalized Brca1F/− MEFs were infected with retroviruses expressing the indicated shRNAs and/or Cre recombinase, followed by selection with puromycin for 72 h. Cells were arrested in mitosis with colcemid and mitotic chromosomes were processed for CO-FISH analysis. Metaphase chromosome spreads were stained with Cy3-conjugated leading strand telomeric PNA probe (red) and FITC-conjugated lagging strand telomeric PNA probe (green). DNA was counter-stained with DAPI (blue).
The frequency of chromosome-type telomeric fusions in cells treated as in (A) was quantified as a percentage of total number of chromosomes (illustrated in Supplementary Fig S2A). A minimum of 2,000 chromosomes were scored for each treatment. Error bars represent SD of at least two independent experiments. P-values were calculated using an unpaired two-tailed t-test. *P ≤ 0.05; **P ≤ 0.01; NS, P > 0.05.
Immortalized Brca1F/− MEFs were infected with retroviruses expressing the indicated shRNAs and/or Cre recombinase, followed by selection with puromycin for 72 h. Mitotic chromosomes isolated 48 h later were fixed and stained with a Cy3-conjugated (CCCTAA)3-PNA probe. The frequency of end-to-end chromosome-type fusions is represented as a percentage of fusions observed after TRF2 depletion. A minimum of 2,000 chromosomes were scored for each sample. Error bars represent SD of three independent experiments. P-values were calculated using an unpaired two-tailed t-test. NS, P > 0.05.
Immortalized Brca1F/C61G MEFs were infected with retroviruses expressing the indicated shRNAs and/or Cre recombinase, followed by selection with puromycin for 72 h. The frequency of end-to-end chromosome-type fusions was analysed as in (A).
MEFs of the indicated genotypes were infected with retroviruses expressing TRF2 and/or CtIP shRNAs, followed by selection with puromycin for 72 h. Cell extracts were prepared 48 h later and analysed by Western blotting as indicated. GAPDH was used as a loading control. *non-specific band. Cells treated as in (C) and (E) were arrested in mitosis with colcemid, and mitotic chromosomes isolated 48 h later were fixed and stained with a Cy3-conjugated (CCCTAA)3-PNA probe (D, F). The frequency of end-to-end chromosome-type fusions is represented as a percentage of fusions observed after TRF2 depletion. Error bars represent SD of two independent experiments. P-values were calculated using an unpaired two-tailed t-test. *P ≤ 0.05.

Immortalized MEFs were infected with retroviruses expressing the indicated shRNAs, followed by selection with puromycin for 72 h. Cell extracts were prepared 48 h later and analysed by Western blotting as indicated. SMC1 was used as a loading control. *non-specific band.
Quantification of the frequency of end-to-end chromosome-type fusions of cells treated as in (A) represented as a percentage of fusions observed after TRF2 depletion. A minimum of 2,000 chromosomes were scored for each sample. Error bars represent SD of three independent experiments. The P-value was calculated using an unpaired two-tailed t-test. NS, P > 0.05.
Immortalized Brca1F/− MEFs were infected with retroviruses expressing the indicated shRNAs and/or Cre recombinase. Mitotic chromosomes isolated 48 h later were fixed and stained with a Cy3-conjugated (CCCTAA)3-PNA probe. The frequency of end-to-end chromosome-type fusions is represented as a percentage of fusions observed after TRF2 depletion. A minimum of 2,000 chromosomes were scored for each sample. Error bars represent SD of three independent experiments. P-values were calculated using an unpaired two-tailed t-test. NS, P > 0.05.
Immortalized Brca1F/− MEFs were infected with retroviruses expressing TRF2 shRNAs and/or Cre recombinase, together with a lentivirus expressing PARP1 shRNA, followed by selection with puromycin for 72 h. Cell extracts were prepared 48 h later and analysed by Western blotting as indicated. SMC1 was used as a loading control. *non-specific band.
Quantification of the frequency of end-to-end chromosome-type fusions of cells treated as in (D) represented as a percentage of fusions observed after TRF2 depletion. A minimum of 1,500 chromosomes were scored for each sample. Error bars represent SD of three independent experiments. The P-value was calculated using an unpaired two-tailed t-test. NS, P > 0.05.
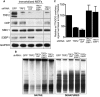
Immortalized MEFs were infected with retroviruses expressing the indicated shRNAs, followed by selection with puromycin for 72 h. Cell extracts were prepared 48 h later and analysed by Western blotting as indicated. SMC1 and GAPDH were used as loading controls. *non-specific band.
MboI- and AluI-digested DNA from cells treated as in (A) was resolved by pulsed-field gel electrophoresis and probed with end-labelled (AACCCT)4 probe. Representative pulsed-field gel samples run under native and denatured conditions are shown.
Quantification of the 3′ overhang in cells treated as in (B). For each sample, the ss/total DNA ratios were expressed relative to the GFP shRNA-treated control. Error bars represent SD of two independent experiments.
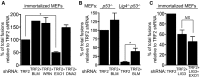
MEFs of the indicated genotypes were infected with retroviruses expressing the indicated shRNAs, followed by selection with puromycin for 72 h. Mitotic chromosomes isolated 48 h later were fixed and stained with a Cy3-conjugated (CCCTAA)3-PNA probe. The frequency of end-to-end chromosome-type fusions is represented as a percentage of fusions observed after TRF2 depletion in each experiment. A minimum of 1,200 chromosomes were scored for each sample. Error bars represent SD of two independent experiments. P-values were calculated using an unpaired two-tailed t-test. *P ≤ 0.05; NS, P > 0.05. Each graph represents a separate set of experiments.

References
-
- Audebert M, Salles B, Calsou P. Involvement of poly(ADP-ribose) polymerase-1 and XRCC1/DNA ligase III in an alternative route for DNA double-strand breaks rejoining. J Biol Chem. 2004;279:55117–55126. - PubMed
-
- Bailey SM, Cornforth MN, Kurimasa A, Chen DJ, Goodwin EH. Strand-specific postreplicative processing of mammalian telomeres. Science. 2001;293:2462–2465. - PubMed
-
- Barnes DE, Stamp G, Rosewell I, Denzel A, Lindahl T. Targeted disruption of the gene encoding DNA ligase IV leads to lethality in embryonic mice. Curr Biol. 1998;8:1395–1398. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

