The cutaneous microbiome in outpatients presenting with acute skin abscesses
- PMID: 25583170
- PMCID: PMC4539909
- DOI: 10.1093/infdis/jiv003
The cutaneous microbiome in outpatients presenting with acute skin abscesses
Abstract
Background: Previous studies have demonstrated an association between antibiotic use and the development of skin abscesses. We tested the hypothesis that alterations in the composition of the cutaneous microbiota may predispose individuals to skin abscesses.
Methods: We studied 25 patients with skin abscesses and 25 age-matched controls, who each completed a questionnaire. Skin swab samples were obtained for DNA analysis from 4 sites around the abscess site (hereafter, "peri-abscess specimens") and from similar sites on the patient's contralateral side and on healthy control subjects. DNA was extracted and analyzed by quantitative polymerase chain reaction (qPCR) and high-throughput sequencing. The purulent abscess drainage was sent for culture.
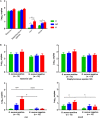
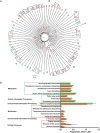
Results: Fifteen patients with abscess were infected with Staphylococcus aureus. Use of nuc qPCR to quantitate S. aureus revealed a significantly greater frequency of positive results for peri-abscess and contralateral skin samples, compared with control skin specimens. Analysis of community structure showed greater heterogeneity in the control samples than in the peri-abscess and contralateral samples. Metagenomic analysis detected significantly more predicted genes related to metabolic activity in the peri-abscess specimens than in the control samples.
Conclusions: The peri-abscess microbiome was similar to the contralateral microbiome, but both microbiomes differed from that for control patients. Host characteristics affecting microbial populations might be important determinants of abscess risk.
Keywords: MRSA; abscess; skin infection; skin microbiome.
© The Author 2015. Published by Oxford University Press on behalf of the Infectious Diseases Society of America. All rights reserved. For Permissions, please e-mail: journals.permissions@oup.com.
Figures


References
-
- Singer AJ, Talan DA. Management of skin abscesses in the era of methicillin-resistant Staphylococcus aureus. N Engl J Med 2014; 370:1031–47. - PubMed
-
- Moran GJ, Krishnadasan A, Gorwitx RJ. Methicillin-resistant S. aureus infections among patients in the emergency department. N Engl J Med 2006; 355:666–74. - PubMed
-
- Sullivan A, Edlund C, Nord CE. Effect of antimicrobial agents on the ecological balance of human microflora. Lancet Infect Dis 2001; 1:101–14. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

