Activating mutations of STAT5B and STAT3 in lymphomas derived from γδ-T or NK cells
- PMID: 25586472
- PMCID: PMC7743911
- DOI: 10.1038/ncomms7025
Activating mutations of STAT5B and STAT3 in lymphomas derived from γδ-T or NK cells
Abstract
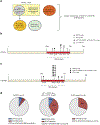
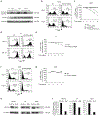
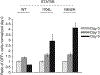
Lymphomas arising from NK or γδ-T cells are very aggressive diseases and little is known regarding their pathogenesis. Here we report frequent activating mutations of STAT3 and STAT5B in NK/T-cell lymphomas (n=51), γδ-T-cell lymphomas (n=43) and their cell lines (n=9) through next generation and/or Sanger sequencing. STAT5B N642H is particularly frequent in all forms of γδ-T-cell lymphomas. STAT3 and STAT5B mutations are associated with increased phosphorylated protein and a growth advantage to transduced cell lines or normal NK cells. Growth-promoting activity of the mutants can be partially inhibited by a JAK1/2 inhibitor. Molecular modelling and surface plasmon resonance measurements of the N642H mutant indicate a marked increase in binding affinity of the phosphotyrosine-Y699 with the mutant histidine. This is associated with the prolonged persistence of the mutant phosphoSTAT5B and marked increase of binding to target sites. Our findings suggest that JAK-STAT pathway inhibition may represent a therapeutic strategy.
Figures




References
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

