Transposition of a rice Mutator-like element in the yeast Saccharomyces cerevisiae
- PMID: 25587002
- PMCID: PMC4330571
- DOI: 10.1105/tpc.114.128488
Transposition of a rice Mutator-like element in the yeast Saccharomyces cerevisiae
Abstract
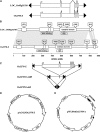
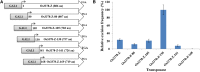
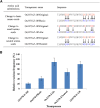
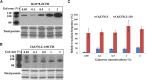
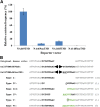
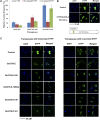
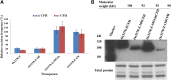
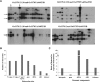
Mutator-like transposable elements (MULEs) are widespread in plants and are well known for their high transposition activity as well as their ability to duplicate and amplify host gene fragments. Despite their abundance and importance, few active MULEs have been identified. In this study, we demonstrated that a rice (Oryza sativa) MULE, Os3378, is capable of excising and reinserting in yeast (Saccharomyces cerevisiae), suggesting that yeast harbors all the host factors for the transposition of MULEs. The transposition activity induced by the wild-type transposase is low but can be altered by modification of the transposase sequence, including deletion, fusion, and substitution. Particularly, fusion of a fluorescent protein to the transposase enhanced the transposition activity, representing another approach to manipulate transposases. Moreover, we identified a critical region in the transposase where the net charge of the amino acids seems to be important for activity. Finally, transposition efficiency is also influenced by the element and its flanking sequences (i.e., small elements are more competent than their large counterparts). Perfect target site duplication is favorable, but not required, for precise excision. In addition to the potential application in functional genomics, this study provides the foundation for further studies of the transposition mechanism of MULEs.
© 2015 American Society of Plant Biologists. All rights reserved.
Figures
Comment in
-
Rice MULEs transpose in yeast.Plant Cell. 2015 Jan;27(1):5-6. doi: 10.1105/tpc.114.136143. Epub 2015 Jan 16. Plant Cell. 2015. PMID: 25596003 Free PMC article. No abstract available.
References
-
- Bachmair A., Finley D., Varshavsky A. (1986). In vivo half-life of a protein is a function of its amino-terminal residue. Science 234: 179–186. - PubMed
-
- Bennetzen J.L. (1996). The Mutator transposable element system of maize. Curr. Top. Microbiol. Immunol. 204: 195–229. - PubMed
-
- Chalvet F., Grimaldi C., Kaper F., Langin T., Daboussi M.J. (2003). Hop, an active Mutator-like element in the genome of the fungus Fusarium oxysporum. Mol. Biol. Evol. 20: 1362–1375. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

