A unifying gene signature for adenoid cystic cancer identifies parallel MYB-dependent and MYB-independent therapeutic targets
- PMID: 25587024
- PMCID: PMC4350357
- DOI: 10.18632/oncotarget.2985
A unifying gene signature for adenoid cystic cancer identifies parallel MYB-dependent and MYB-independent therapeutic targets
Abstract
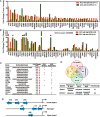
MYB activation is proposed to underlie development of adenoid cystic cancer (ACC), an aggressive salivary gland tumor with no effective systemic treatments. To discover druggable targets for ACC, we performed global mRNA/miRNA analyses of 12 ACC with matched normal tissues, and compared these data with 14 mucoepidermoid carcinomas (MEC) and 11 salivary adenocarcinomas (ADC). We detected a unique ACC gene signature of 1160 mRNAs and 22 miRNAs. MYB was the top-scoring gene (18-fold induction), however we observed the same signature in ACC without detectable MYB gene rearrangements. We also found 4 ACC tumors (1 among our 12 cases and 3 from public databases) with negligible MYB expression that retained the same ACC mRNA signature including over-expression of extracellular matrix (ECM) genes. Integration of this signature with somatic mutational analyses suggests that NOTCH1 and RUNX1 participate with MYB to activate ECM elements including the VCAN/HAPLN1 complex. We observed that forced MYB-NFIB expression in human salivary gland cells alters cell morphology and cell adhesion in vitro and depletion of VCAN blocked tumor cell growth of a short-term ACC tumor culture. In summary, we identified a unique ACC signature with parallel MYB-dependent and independent biomarkers and identified VCAN/HAPLN1 complexes as a potential target.
Figures





Similar articles
-
Clinically significant copy number alterations and complex rearrangements of MYB and NFIB in head and neck adenoid cystic carcinoma.Genes Chromosomes Cancer. 2012 Aug;51(8):805-17. doi: 10.1002/gcc.21965. Epub 2012 Apr 16. Genes Chromosomes Cancer. 2012. PMID: 22505352
-
Adenoid cystic carcinoma of the lacrimal gland: MYB gene activation, genomic imbalances, and clinical characteristics.Ophthalmology. 2013 Oct;120(10):2130-8. doi: 10.1016/j.ophtha.2013.03.030. Epub 2013 May 29. Ophthalmology. 2013. PMID: 23725736
-
Study of MYB-NFIB chimeric gene expression, tumor angiogenesis, and proliferation in adenoid cystic carcinoma of salivary gland.Odontology. 2018 Jul;106(3):238-244. doi: 10.1007/s10266-017-0326-1. Epub 2017 Dec 14. Odontology. 2018. PMID: 29243184
-
New developments in the molecular pathogenesis of head and neck tumors: a review of tumor-specific fusion oncogenes in mucoepidermoid carcinoma, adenoid cystic carcinoma, and NUT midline carcinoma.Ann Diagn Pathol. 2011 Feb;15(1):69-77. doi: 10.1016/j.anndiagpath.2010.12.001. Ann Diagn Pathol. 2011. PMID: 21238915 Review.
-
MYB-NFIB fusion transcript in adenoid cystic carcinoma: Current state of knowledge and future directions.Crit Rev Oncol Hematol. 2022 Aug;176:103745. doi: 10.1016/j.critrevonc.2022.103745. Epub 2022 Jun 20. Crit Rev Oncol Hematol. 2022. PMID: 35738530 Review.
Cited by
-
Genetic Rearrangements in Different Salivary Gland Tumors: A Systematic Review.Cureus. 2024 Jun 4;16(6):e61639. doi: 10.7759/cureus.61639. eCollection 2024 Jun. Cureus. 2024. PMID: 38966479 Free PMC article. Review.
-
Pulmonary adenoid cystic carcinoma: molecular characteristics and literature review.Diagn Pathol. 2023 May 17;18(1):65. doi: 10.1186/s13000-023-01354-4. Diagn Pathol. 2023. PMID: 37198615 Free PMC article. Review.
-
Identification of β-catenin target genes in colorectal cancer by interrogating gene fitness screening data.Oncol Lett. 2019 Oct;18(4):3769-3777. doi: 10.3892/ol.2019.10724. Epub 2019 Aug 6. Oncol Lett. 2019. PMID: 31516589 Free PMC article.
-
MYB-QKI rearrangements in angiocentric glioma drive tumorigenicity through a tripartite mechanism.Nat Genet. 2016 Mar;48(3):273-82. doi: 10.1038/ng.3500. Epub 2016 Feb 1. Nat Genet. 2016. PMID: 26829751 Free PMC article.
-
Whole-Genome Sequencing of Salivary Gland Adenoid Cystic Carcinoma.Cancer Prev Res (Phila). 2016 Apr;9(4):265-74. doi: 10.1158/1940-6207.CAPR-15-0316. Epub 2016 Feb 9. Cancer Prev Res (Phila). 2016. PMID: 26862087 Free PMC article.
References
-
- L Barnes JE, Reichart P, Sidransky D. Pathology and Genetics of Head and Neck Tumors. Lyon: IARC Press; 2005. World Health Organization International Classification of Tumors.
-
- Tonon G, Modi S, Wu L, Kubo A, Coxon AB, Komiya T, O'Neil K, Stover K, El-Naggar A, Griffin JD, Kirsch IR, Kaye FJ. t(11;19)(q21;p13) translocation in mucoepidermoid carcinoma creates a novel fusion product that disrupts a Notch signaling pathway. Nat Genet. 2003;33(2):208–213. - PubMed
-
- Tirado Y, Williams MD, Hanna EY, Kaye FJ, Batsakis JG, El-Naggar AK. CRTC1/MAML2 fusion transcript in high grade mucoepidermoid carcinomas of salivary and thyroid glands and Warthin's tumors: implications for histogenesis and biologic behavior. Genes Chromosomes Cancer. 2007;46(7):708–715. - PubMed
-
- Coxon A, Rozenblum E, Park YS, Joshi N, Tsurutani J, Dennis PA, Kirsch IR, Kaye FJ. Mect1-Maml2 fusion oncogene linked to the aberrant activation of cyclic AMP/CREB regulated genes. Cancer Res. 2005;65(16):7137–7144. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

