MAPK feedback encodes a switch and timer for tunable stress adaptation in yeast
- PMID: 25587192
- PMCID: PMC4505820
- DOI: 10.1126/scisignal.2005774
MAPK feedback encodes a switch and timer for tunable stress adaptation in yeast
Abstract
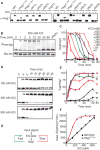
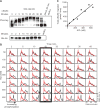
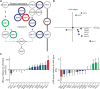
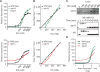
Signaling pathways can behave as switches or rheostats, generating binary or graded responses to a given cell stimulus. We evaluated whether a single signaling pathway can simultaneously encode a switch and a rheostat. We found that the kinase Hog1 mediated a bifurcated cellular response: Activation and commitment to adaptation to osmotic stress are switchlike, whereas protein induction and the resolution of this commitment are graded. Through experimentation, bioinformatics analysis, and computational modeling, we determined that graded recovery is encoded through feedback phosphorylation and a gene induction program that is both temporally staggered and variable across the population. This switch-to-rheostat signaling mechanism represents a versatile stress adaptation system, wherein a broad range of inputs generate an "all-in" response that is later tuned to allow graded recovery of individual cells over time.
Copyright © 2015, American Association for the Advancement of Science.
Conflict of interest statement
Competing interests: The authors declare that they have no competing interests.
Figures



References
-
- Wagner EF, Nebreda ÁR. Signal integration by JNK and p38 MAPK pathways in cancer development. Nat Rev Cancer. 2009;9:537–549. - PubMed
-
- Brewster JL, de Valoir T, Dwyer ND, Winter E, Gustin MC. An osmosensing signal transduction pathway in yeast. Science. 1993;259:1760–1763. - PubMed
-
- Rouse J, Cohen P, Trigon S, Morange M, Alonso-Llamazares A, Zamanillo D, Hunt T, Nebreda AR. A novel kinase cascade triggered by stress and heat shock that stimulates MAPKAP kinase-2 and phosphorylation of the small heat shock proteins. Cell. 1994;78:1027–1037. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

