Transcription activator-like effector nuclease (TALEN)-mediated CLYBL targeting enables enhanced transgene expression and one-step generation of dual reporter human induced pluripotent stem cell (iPSC) and neural stem cell (NSC) lines
- PMID: 25587899
- PMCID: PMC4294658
- DOI: 10.1371/journal.pone.0116032
Transcription activator-like effector nuclease (TALEN)-mediated CLYBL targeting enables enhanced transgene expression and one-step generation of dual reporter human induced pluripotent stem cell (iPSC) and neural stem cell (NSC) lines
Abstract
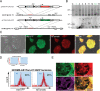
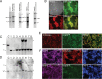
Targeted genome engineering to robustly express transgenes is an essential methodology for stem cell-based research and therapy. Although designer nucleases have been used to drastically enhance gene editing efficiency, targeted addition and stable expression of transgenes to date is limited at single gene/locus and mostly PPP1R12C/AAVS1 in human stem cells. Here we constructed transcription activator-like effector nucleases (TALENs) targeting the safe-harbor like gene CLYBL to mediate reporter gene integration at 38%-58% efficiency, and used both AAVS1-TALENs and CLYBL-TALENs to simultaneously knock-in multiple reporter genes at dual safe-harbor loci in human induced pluripotent stem cells (iPSCs) and neural stem cells (NSCs). The CLYBL-TALEN engineered cell lines maintained robust reporter expression during self-renewal and differentiation, and revealed that CLYBL targeting resulted in stronger transgene expression and less perturbation on local gene expression than PPP1R12C/AAVS1. TALEN-mediated CLYBL engineering provides improved transgene expression and options for multiple genetic modification in human stem cells.
Conflict of interest statement
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

