Defining the phylogenomics of Shigella species: a pathway to diagnostics
- PMID: 25588655
- PMCID: PMC4390639
- DOI: 10.1128/JCM.03527-14
Defining the phylogenomics of Shigella species: a pathway to diagnostics
Abstract
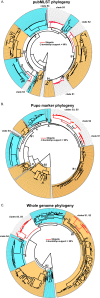
Shigellae cause significant diarrheal disease and mortality in humans, as there are approximately 163 million episodes of shigellosis and 1.1 million deaths annually. While significant strides have been made in the understanding of the pathogenesis, few studies on the genomic content of the Shigella species have been completed. The goal of this study was to characterize the genomic diversity of Shigella species through sequencing of 55 isolates representing members of each of the four Shigella species: S. flexneri, S. sonnei, S. boydii, and S. dysenteriae. Phylogeny inferred from 336 available Shigella and Escherichia coli genomes defined exclusive clades of Shigella; conserved genomic markers that can identify each clade were then identified. PCR assays were developed for each clade-specific marker, which was combined with an amplicon for the conserved Shigella invasion antigen, IpaH3, into a multiplex PCR assay. This assay demonstrated high specificity, correctly identifying 218 of 221 presumptive Shigella isolates, and sensitivity, by not identifying any of 151 diverse E. coli isolates incorrectly as Shigella. This new phylogenomics-based PCR assay represents a valuable tool for rapid typing of uncharacterized Shigella isolates and provides a framework that can be utilized for the identification of novel genomic markers from genomic data.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures


References
-
- Niyogi SK. 2005. Shigellosis. J Microbiol 43:133–143. - PubMed
-
- WHO. 2009, posting date Initiative for vaccine research (IVR): diarrhoeal diseases. WHO, Geneva, Switzerland.
-
- Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, Panchalingam S, Wu Y, Sow SO, Sur D, Breiman RF, Faruque AS, Zaidi AK, Saha D, Alonso PL, Tamboura B, Sanogo D, Onwuchekwa U, Manna B, Ramamurthy T, Kanungo S, Ochieng JB, Omore R, Oundo JO, Hossain A, Das SK, Ahmed S, Qureshi S, Quadri F, Adegbola RA, Antonio M, Hossain MJ, Akinsola A, Mandomando I, Nhampossa T, Acacio S, Biswas K, O'Reilly CE, Mintz ED, Berkeley LY, Muhsen K, Sommerfelt H, Robins-Browne RM, Levine MM. 2013. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet 382:209–222. doi:10.1016/S0140-6736(13)60844-2. - DOI - PubMed
-
- Kotloff KL, Blackwelder WC, Nasrin D, Nataro JP, Farag TH, van Eijk A, Adegbola RA, Alonso PL, Breiman RF, Faruque AS, Saha D, Sow SO, Sur D, Zaidi AK, Biswas K, Panchalingam S, Clemens JD, Cohen D, Glass RI, Mintz ED, Sommerfelt H, Levine MM. 2012. The Global Enteric Multicenter Study (GEMS) of diarrheal disease in infants and young children in developing countries: epidemiologic and clinical methods of the case/control study. Clin Infect Dis 55(Suppl 4):S232–S245. doi:10.1093/cid/cis753. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

