Temporal and evolutionary dynamics of two-component signaling pathways
- PMID: 25589045
- PMCID: PMC4380680
- DOI: 10.1016/j.mib.2014.12.003
Temporal and evolutionary dynamics of two-component signaling pathways
Abstract
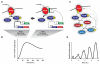
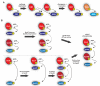
Bacteria sense and respond to numerous environmental signals through two-component signaling pathways. Typically, a given stimulus will activate a sensor histidine kinase to autophosphorylate and then phosphotransfer to a cognate response regulator, which can mount an appropriate response. Although these signaling pathways often appear to be simple switches, they can also orchestrate surprisingly sophisticated and complex responses. These temporal dynamics arise from several key regulatory features, including the bifunctionality of histidine kinases as well as positive and negative feedback loops. Two-component signaling pathways are also dynamic on evolutionary time-scales, expanding dramatically in many species through gene duplication and divergence. Here, we review recent work probing the temporal and evolutionary dynamics of two-component signaling systems.
Copyright © 2015 Elsevier Ltd. All rights reserved.
Figures
Similar articles
-
Histidine kinases and response regulators in networks.Curr Opin Microbiol. 2012 Apr;15(2):118-24. doi: 10.1016/j.mib.2011.11.009. Epub 2011 Dec 13. Curr Opin Microbiol. 2012. PMID: 22172627 Review.
-
Nitric oxide regulated two-component signaling in Pseudoalteromonas atlantica.Biochem Biophys Res Commun. 2012 May 11;421(3):521-6. doi: 10.1016/j.bbrc.2012.04.037. Epub 2012 Apr 10. Biochem Biophys Res Commun. 2012. PMID: 22521885
-
The Histidine Residue of QseC Is Required for Canonical Signaling between QseB and PmrB in Uropathogenic Escherichia coli.J Bacteriol. 2017 Aug 22;199(18):e00060-17. doi: 10.1128/JB.00060-17. Print 2017 Sep 15. J Bacteriol. 2017. PMID: 28396353 Free PMC article.
-
Spatial tethering of kinases to their substrates relaxes evolutionary constraints on specificity.Mol Microbiol. 2012 Dec;86(6):1393-403. doi: 10.1111/mmi.12064. Epub 2012 Nov 5. Mol Microbiol. 2012. PMID: 23078131
-
Tearing down the wall: peptidoglycan metabolism and the WalK/WalR (YycG/YycF) essential two-component system.Adv Exp Med Biol. 2008;631:214-28. doi: 10.1007/978-0-387-78885-2_15. Adv Exp Med Biol. 2008. PMID: 18792692 Review.
Cited by
-
Quantitative Kinetic Analyses of Shutting Off a Two-Component System.mBio. 2017 May 16;8(3):e00412-17. doi: 10.1128/mBio.00412-17. mBio. 2017. PMID: 28512092 Free PMC article.
-
Comparison of the antibiotic resistance mechanisms in a gram-positive and a gram-negative bacterium by gene networks analysis.PLoS One. 2024 Nov 15;19(11):e0311434. doi: 10.1371/journal.pone.0311434. eCollection 2024. PLoS One. 2024. PMID: 39546505 Free PMC article.
-
Proteolysis of histidine kinase VgrS inhibits its autophosphorylation and promotes osmostress resistance in Xanthomonas campestris.Nat Commun. 2018 Nov 15;9(1):4791. doi: 10.1038/s41467-018-07228-4. Nat Commun. 2018. PMID: 30442885 Free PMC article.
-
An experimentally supported model of the Bacillus subtilis global transcriptional regulatory network.Mol Syst Biol. 2015 Nov 17;11(11):839. doi: 10.15252/msb.20156236. Mol Syst Biol. 2015. PMID: 26577401 Free PMC article.
-
Increasing intracellular magnesium levels with the 31-amino acid MgtS protein.Proc Natl Acad Sci U S A. 2017 May 30;114(22):5689-5694. doi: 10.1073/pnas.1703415114. Epub 2017 May 16. Proc Natl Acad Sci U S A. 2017. PMID: 28512220 Free PMC article.
References
-
- Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu. Rev. Biochem. 2000;69:183–215. - PubMed
-
- Krell T, Lacal J, Busch A, Silva-Jiménez H, Guazzaroni M-E, Ramos JL. Bacterial Sensor Kinases: Diversity in the Recognition of Environmental Signals. Annu. Rev. Microbiol. 2010;64:539–559. - PubMed
-
- Podgornaia AI, Laub MT. Determinants of specificity in two-component signal transduction. Curr. Opin. Microbiol. 2013;16:156–162. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

