The Release 6 reference sequence of the Drosophila melanogaster genome
- PMID: 25589440
- PMCID: PMC4352887
- DOI: 10.1101/gr.185579.114
The Release 6 reference sequence of the Drosophila melanogaster genome
Abstract
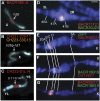
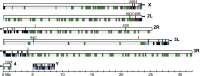
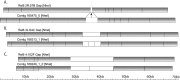
Drosophila melanogaster plays an important role in molecular, genetic, and genomic studies of heredity, development, metabolism, behavior, and human disease. The initial reference genome sequence reported more than a decade ago had a profound impact on progress in Drosophila research, and improving the accuracy and completeness of this sequence continues to be important to further progress. We previously described improvement of the 117-Mb sequence in the euchromatic portion of the genome and 21 Mb in the heterochromatic portion, using a whole-genome shotgun assembly, BAC physical mapping, and clone-based finishing. Here, we report an improved reference sequence of the single-copy and middle-repetitive regions of the genome, produced using cytogenetic mapping to mitotic and polytene chromosomes, clone-based finishing and BAC fingerprint verification, ordering of scaffolds by alignment to cDNA sequences, incorporation of other map and sequence data, and validation by whole-genome optical restriction mapping. These data substantially improve the accuracy and completeness of the reference sequence and the order and orientation of sequence scaffolds into chromosome arm assemblies. Representation of the Y chromosome and other heterochromatic regions is particularly improved. The new 143.9-Mb reference sequence, designated Release 6, effectively exhausts clone-based technologies for mapping and sequencing. Highly repeat-rich regions, including large satellite blocks and functional elements such as the ribosomal RNA genes and the centromeres, are largely inaccessible to current sequencing and assembly methods and remain poorly represented. Further significant improvements will require sequencing technologies that do not depend on molecular cloning and that produce very long reads.
© 2015 Hoskins et al.; Published by Cold Spring Harbor Laboratory Press.
Figures

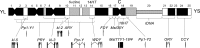
References
-
- Abad JP, Agudo M, Molina I, Losada A, Ripoll P, Villasante A. 2000. Pericentromeric regions containing 1.688 satellite DNA sequences show anti-kinetochore antibody staining in prometaphase chromosomes of Drosophila melanogaster. Mol Gen Genet 264: 371–377. - PubMed
-
- Abad JP, de Pablos B, Agudo M, Molina I, Giovinazzo G, Martin-Gallardo A, Villasante A. 2004a. Genomic and cytological analysis of the Y chromosome of Drosophila melanogaster: telomere-derived sequences at internal regions. Chromosoma 113: 295–304. - PubMed
-
- Abad JP, De Pablos B, Osoegawa K, De Jong PJ, Martin-Gallardo A, Villasante A. 2004b. Genomic analysis of Drosophila melanogaster telomeres: full-length copies of HeT-A and TART elements at telomeres. Mol Biol Evol 21: 1613–1619. - PubMed
-
- Accardo MC, Dimitri P. 2010. Fluorescence in situ hybridization with Bacterial Artificial Chromosomes (BACs) to mitotic heterochromatin of Drosophila. Methods Mol Biol 659: 389–400. - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
