DNA lesion identity drives choice of damage tolerance pathway in murine cell chromosomes
- PMID: 25589543
- PMCID: PMC4330363
- DOI: 10.1093/nar/gku1398
DNA lesion identity drives choice of damage tolerance pathway in murine cell chromosomes
Abstract
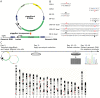
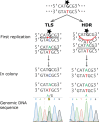
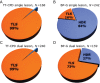
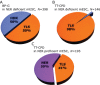
DNA-damage tolerance (DDT) via translesion DNA synthesis (TLS) or homology-dependent repair (HDR) functions to bypass DNA lesions encountered during replication, and is critical for maintaining genome stability. Here, we present piggyBlock, a new chromosomal assay that, using piggyBac transposition of DNA containing a known lesion, measures the division of labor between the two DDT pathways. We show that in the absence of DNA damage response, tolerance of the most common sunlight-induced DNA lesion, TT-CPD, is achieved by TLS in mouse embryo fibroblasts. Meanwhile, BP-G, a major smoke-induced DNA lesion, is bypassed primarily by HDR, providing the first evidence for this mechanism being the main tolerance pathway for a biologically important lesion in a mammalian genome. We also show that, far from being a last-resort strategy as it is sometimes portrayed, TLS operates alongside nucleotide excision repair, handling 40% of TT-CPDs in repair-proficient cells. Finally, DDT acts in mouse embryonic stem cells, exhibiting the same pattern—mutagenic TLS included—despite the risk of propagating mutations along all cell lineages. The new method highlights the importance of HDR, and provides an effective tool for studying DDT in mammalian cells.
Figures
References
-
- Friedberg E.C. DNA Repair And Mutagenesis. Herndon, VA: Amer Society for Microbiology; 2006.
-
- Friedberg E.C. Suffering in silence: the tolerance of DNA damage. Nat. Rev. Mol. Cell. Biol. 2005;6:943–953. - PubMed
-
- Lehmann A.R., Fuchs R.P. Gaps and forks in DNA replication: rediscovering old models. DNA Repair. 2006;5:1495–1498. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

