A phylogenetic backbone for Bivalvia: an RNA-seq approach
- PMID: 25589608
- PMCID: PMC4308999
- DOI: 10.1098/rspb.2014.2332
A phylogenetic backbone for Bivalvia: an RNA-seq approach
Abstract
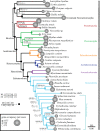
Bivalves are an ancient and ubiquitous group of aquatic invertebrates with an estimated 10 000-20 000 living species. They are economically significant as a human food source, and ecologically important given their biomass and effects on communities. Their phylogenetic relationships have been studied for decades, and their unparalleled fossil record extends from the Cambrian to the Recent. Nevertheless, a robustly supported phylogeny of the deepest nodes, needed to fully exploit the bivalves as a model for testing macroevolutionary theories, is lacking. Here, we present the first phylogenomic approach for this important group of molluscs, including novel transcriptomic data for 31 bivalves obtained through an RNA-seq approach, and analyse these data with published genomes and transcriptomes of other bivalves plus outgroups. Our results provide a well-resolved, robust phylogenetic backbone for Bivalvia with all major lineages delineated, addressing long-standing questions about the monophyly of Protobranchia and Heterodonta, and resolving the position of particular groups such as Palaeoheterodonta, Archiheterodonta and Anomalodesmata. This now fully resolved backbone demonstrates that genomic approaches using hundreds of genes are feasible for resolving phylogenetic questions in bivalves and other animals.
Keywords: bivalves; mollusca; phylogenetics; phylogenomics.
Figures


References
-
- Pawiro S. 2010. Bivalves: global production and trade trends. In Safe management of shellfish and harvest waters (eds Rees G, Pond K, Kay D, Bartram J, Santo Domingo J.), pp. 11–19. London, UK: IWA Publishing.
-
- Gage JD, Tyler PA. 1991. Deep-sea biology: a natural history of organisms at the deep-sea floor. Cambridge, UK: Cambridge University Press.
-
- Purchon RD. 1959. Phylogenetic classification of the Lamellibranchia, with special reference to the Protobranchia. Proc. Malacol. Soc. 33, 224–230.
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
