Characterization of Plasmodium ovale curtisi and P. ovale wallikeri in Western Kenya utilizing a novel species-specific real-time PCR assay
- PMID: 25590587
- PMCID: PMC4295880
- DOI: 10.1371/journal.pntd.0003469
Characterization of Plasmodium ovale curtisi and P. ovale wallikeri in Western Kenya utilizing a novel species-specific real-time PCR assay
Abstract
Background: Plasmodium ovale is comprised of two genetically distinct subspecies, P. ovale curtisi and P. ovale wallikeri. Although P. ovale subspecies are similar based on morphology and geographical distribution, allelic differences indicate that P. ovale curtisi and P. ovale wallikeri are genetically divergent. Additionally, potential clinical and latency duration differences between P. ovale curtisi and P. ovale wallikeri demonstrate the need for investigation into the contribution of this neglected malaria parasite to the global malaria burden.
Methods: In order to detect all P. ovale subspecies simultaneously, we developed an inclusive P. ovale-specific real-time PCR assay based on conserved regions between P. ovale curtisi and P. ovale wallikeri in the reticulocyte binding protein 2 (rbp2) gene. Additionally, we characterized the P. ovale subspecies prevalence from 22 asymptomatic malaria infections using multilocus genotyping to discriminate P. ovale curtisi and P. ovale wallikeri.
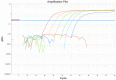
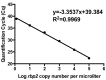
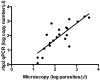
Results: Our P. ovale rbp2 qPCR assay validation experiments demonstrated a linear dynamic range from 6.25 rbp2 plasmid copies/microliter to 100,000 rbp2 plasmid copies/microliter and a limit of detection of 1.5 rbp2 plasmid copies/microliter. Specificity experiments showed the ability of the rbp2 qPCR assay to detect low-levels of P. ovale in the presence of additional malaria parasite species, including P. falciparum, P. vivax, and P. malariae. We identified P. ovale curtisi and P. ovale wallikeri in Western Kenya by DNA sequencing of the tryptophan-rich antigen gene, the small subunit ribosomal RNA gene, and the rbp2 gene.
Conclusions: Our novel P. ovale rbp2 qPCR assay detects P. ovale curtisi and P. ovale wallikeri simultaneously and can be utilized to characterize the prevalence, distribution, and burden of P. ovale in malaria endemic regions. Using multilocus genotyping, we also provided the first description of the prevalence of P. ovale curtisi and P. ovale wallikeri in Western Kenya, a region holoendemic for malaria transmission.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Stevens JWW (1922) A New Malaria Parasite of Man. Ann Trop Med Parasitol 16: 383–388.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

